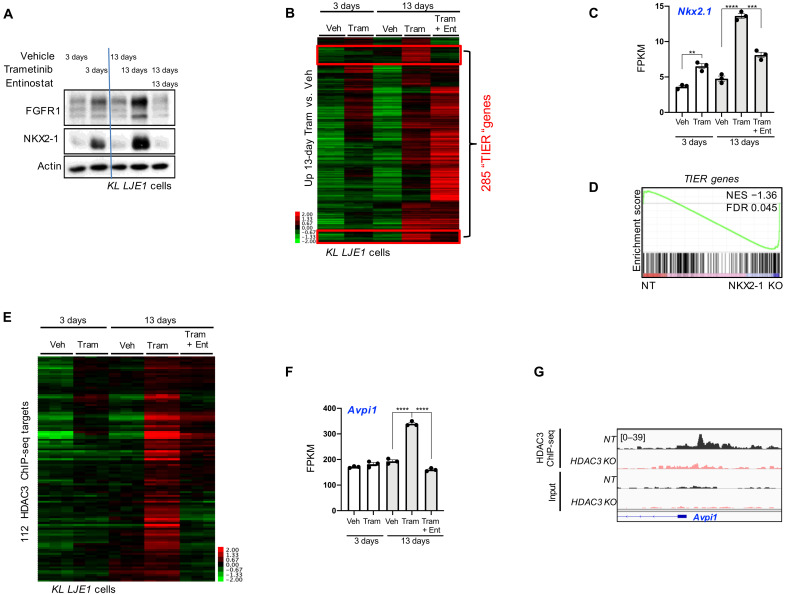
Fig. 4. HDAC3 and NKX2-1 common target genes are aberrantly engaged upon trametinib resistance.
(A) Western blot analysis of protein lysates from KL LJE1 cells treated with vehicle, 10 nM trametinib, or 1 μM entinostat for 3 or 13 days. (B) Heatmap of RNA-seq data showing FPKM read counts across all treatment conditions for the 2141 genes significantly up-regulated (adjusted P < 0.05; fold, >±0.5) upon 13 day of trametinib compared to 13 days of vehicle in KL LJE1 cells. Veh, vehicle; Tram, trametinib; Ent, entinostat. Red boxes identify TIER genes. (C) Nkx2-1 mRNA levels (FPKM) across all treatment conditions (n = 3) from RNA-seq data in (B). (D) Gene set enrichment analysis (GSEA) of the 285 TIER genes queried across RNA-seq data from NKX2-1 KO versus NT KL LJE1 cells. (E) Heatmap of RNA-seq data showing FPKM read counts across all treatment conditions for the 112 TIER genes that are HDAC3 ChIP-seq target genes. (F) Avpi1 mRNA levels across all treatment conditions from RNA-seq data from cells in (B) (n = 3). (G) HDAC3 ChIP-seq data in NT and HDAC3 KO KL LJE1 cells at the Avpi1 genomic locus. Values are expressed as means ± SEM. **P < 0.01, ***P < 0.001, and ****P < 0.0001, determined by two-tailed Student’s t test.