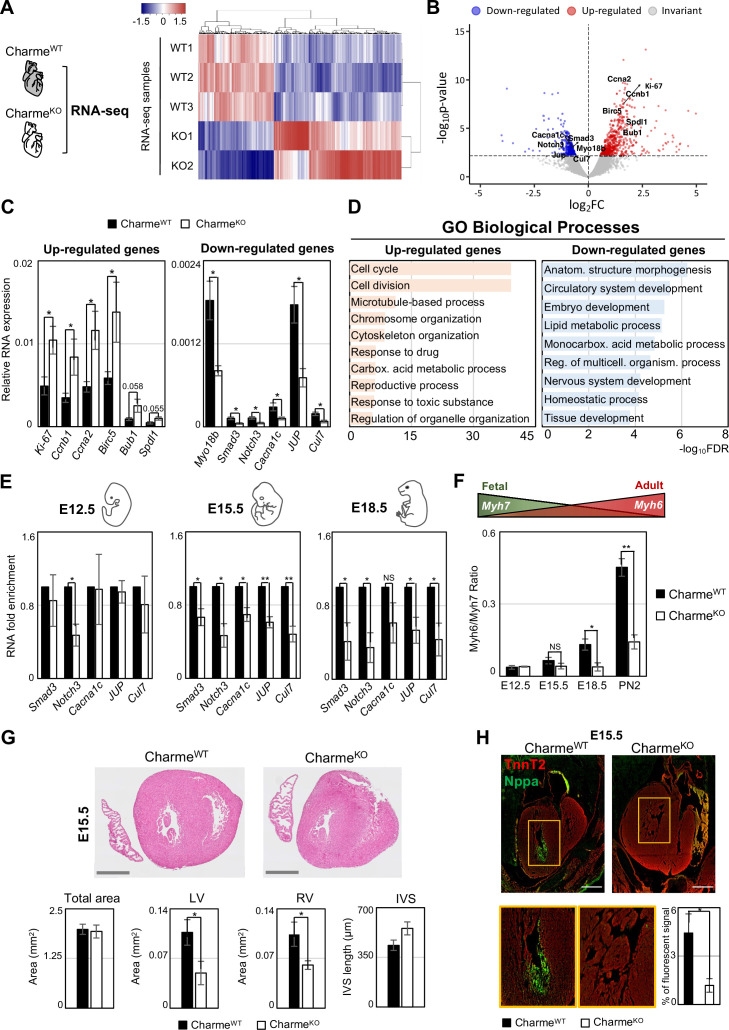
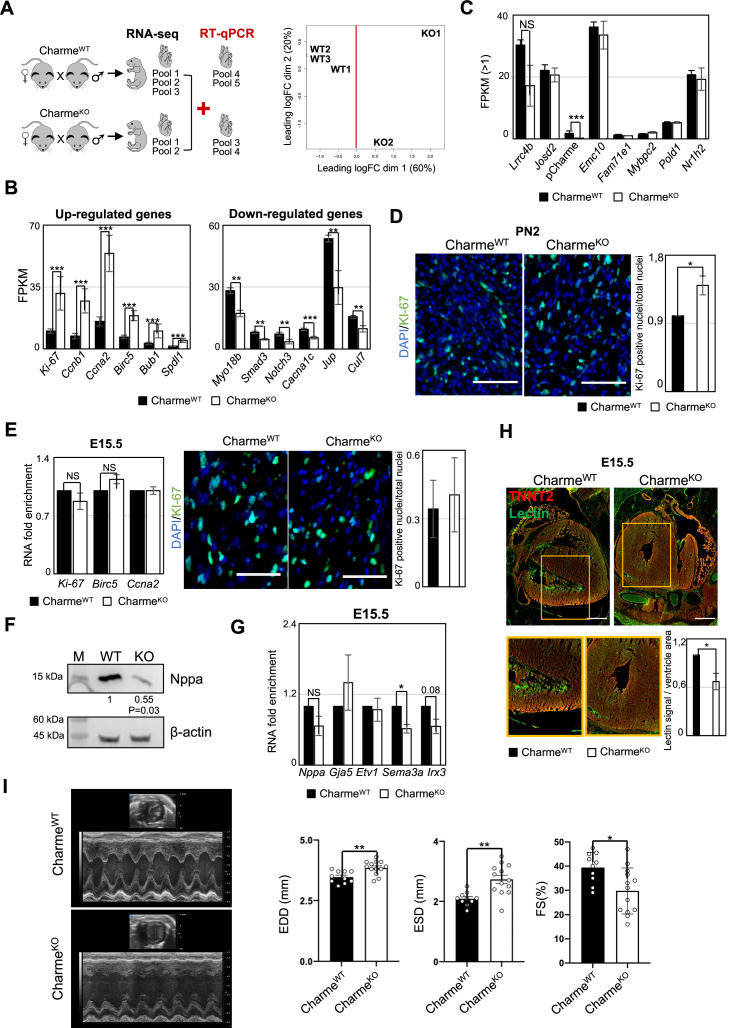
Figure 2. Genome-wide profiling of cardiac CharmeWT and CharmeKO transcriptomes.
(A) Heatmap visualization from RNA-seq analysis of CharmeWT and CharmeKO neonatal (PN2) hearts. Plot was produced by heatmap3 (Zhao, 2021). Expression values were calculated as FPKM, were log2-transformed and mean-centered. FPKM, fragments per kilobase of transcript per million mapped reads. (B) Volcano plots showing differential gene expression from transcriptome analysis of CharmeWT vs. CharmeKO PN2 hearts. Differentially expressed genes (DEGs) validated through RT-qPCR (C) are in evidence. FC, fold change. (C) RT-qPCR quantification of upregulated (left panel) and downregulated (right panel) DEGs in CharmeWT vs. CharmeKO neonatal hearts. Data were normalized to Gapdh mRNA and represent means ± SEM of WT (n = 5) vs. KO (n = 4) independent biological pools (at least 3 littermates/pool). (D) Gene Ontology (GO) enrichment analysis performed by WebGestalt (http://www.webgestalt.org) on upregulated (left panel) and downregulated (right panel) DEGs in CharmeWT vs. CharmeKO pools of neonatal hearts. Bars indicate the top categories of biological processes in decreasing order of – log10FDR. All the represented categories show a false discovery rate (FDR) value <0.05. (E) RT-qPCR quantification of pCharme targets in CharmeWT and CharmeKO extracts from E12.5, E15.5, and E18.5 hearts. DEGs belonging to the GO category ‘anatomical structure morphogenesis’ were considered for the analysis. Data were normalized to Gapdh mRNA and represent means ± SEM of WT and KO (n = 3) independent biological pools (at least 3 littermates/pool). (F) RT-qPCR quantification of the Myh6/Myh7 ratio in CharmeWT and CharmeKO extracts from E12.5, E15.5, and E18.5 and neonatal hearts. Data were normalized to Gapdh mRNA and represent means ± SEM of WT and KO (n = 3) independent biological pools (at least 3 littermates/pool). Schematic representation of the physiological Myh6/Myh7 expression trend is shown. (G) Upper panel: hematoxylin-eosin staining from CharmeWT and CharmeKO E15.5 cardiac transverse sections. Scale bars: 500 μm. Lower panel: quantification of the total area, the left and right ventricle cavities, and the thickness of the interventricular septum (IVS) in CharmeWT and CharmeKO E15.5 hearts. For each genotype, data represent the mean ± SEM of WT and KO (n = 3) biological replicates. (H) Representative images of Nppa (green) and TnnT2 (red) immunostaining in CharmeWT and CharmeKO (E15.5) cardiac sections. Regions of interest (ROI, orange squares) were digitally enlarged on the lower panels. Scale bar: 500 µm. Quantification of the area covered by the Nppa fluorescent signal is shown aside. Data represent the mean (%) ± SEM of WT (n = 4) and KO (n = 3) biological replicates. Data information: *p<0.05; **p<0.01, NS > 0.05, unpaired Student’s t-test.