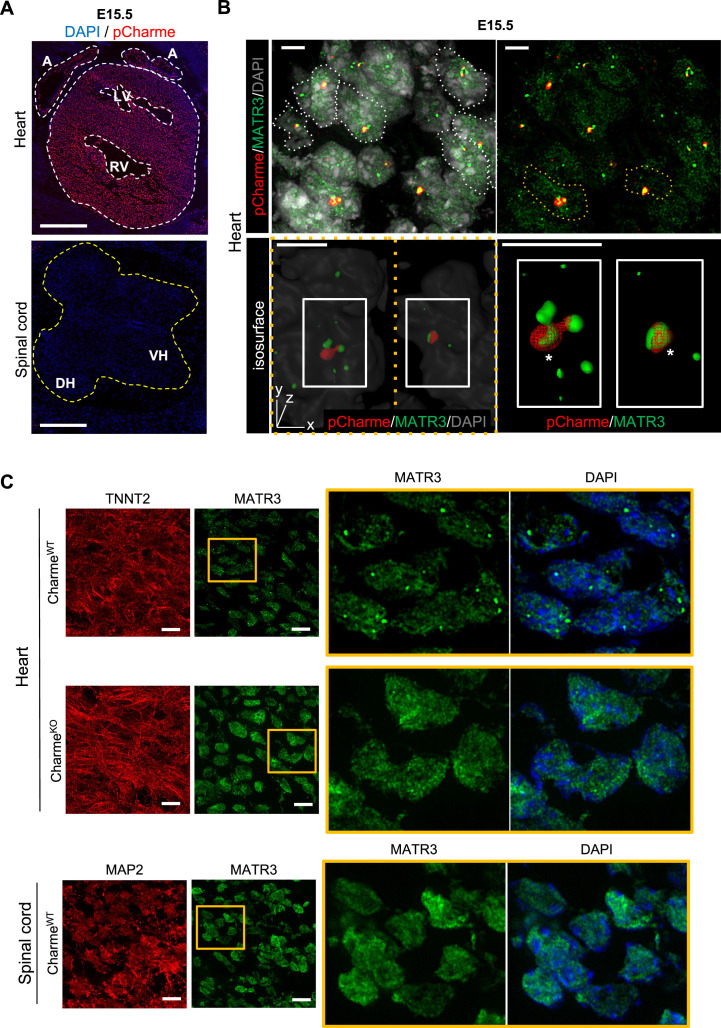
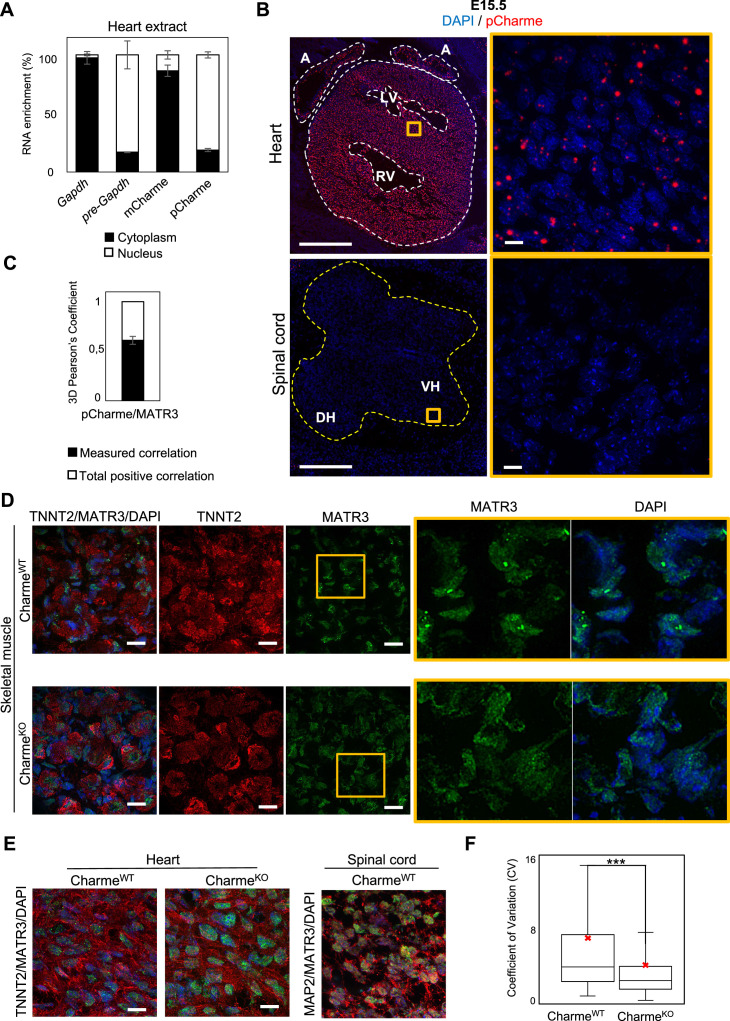
Figure 3. In fetal cardiomyocytes pCharme promotes MATR3 nuclear condensation.
(A) RNA-FISH for pCharme (red) and DAPI staining (blue) in CharmeWT cardiac and spinal cord from E15.5 tissue sections. Whole heart (white dashed lines), spinal cord (yellow dashed line). A, atria; LV and RV, left and right ventricle; DH and VH, dorsal and ventral horn. Scale bars, 500 μm. (B) Upper panel: RNA-FISH for pCharme (red) combined with immunofluorescence for MATR3 (green) and DAPI staining (gray) in CharmeWT from E15.5 cardiac sections. Dashed lines show the edge of nuclei. Lower panel: selected nuclei (yellow dashed lines in the upper panel) were enlarged and processed for isosurface reconstruction (left panel) and digital magnification (right panel). Overlapped signals are shown (asterisks). Scale bars, 5 μm. (C) Upper panel: representative images for MATR3 (green), TnnT2 (red), and DAPI (blue) stainings on CharmeWT and CharmeKO E15.5 cardiac sections. Lower panel: representative images for MATR3 (green), the Microtubule-associated protein 2 (MAP2) (red), and DAPI (blue) stainings on CharmeWT and CharmeKO E15.5 spinal cord sections. Regions of interest (ROI) (orange squares) were digitally enlarged on the right panels. Each image is representative of three individual biological replicates. Scale bars, 10 μm.