Abstract
Hundreds of millions of people worldwide have recently been infected by the novel Coronavirus disease (COVID-19), causing significant damage to the health, economy, and welfare of the world's population. Moreover, the unprecedented number of patients with COVID-19 has placed a massive burden on healthcare centers, making timely and rapid diagnosis challenging. A crucial step in minimizing the impact of such problems is to automatically detect infected patients and place them under special care as quickly as possible. Deep learning algorithms, such as Convolutional Neural Networks (CNN), can be used to meet this need. Despite the desired results, most of the existing deep learning-based models were built on millions of parameters (weights), which are not applicable to devices with limited resources. Inspired by such fact, in this research, we developed two new lightweight CNN-based diagnostic models for the automatic and early detection of COVID-19 subjects from chest X-ray images. The first model was built for binary classification (COVID-19 and Normal), whereas the second one was built for multiclass classification (COVID-19, viral pneumonia, or normal). The proposed models were tested on a relatively large dataset of chest X-ray images, and the results showed that the accuracy rates of the 2- and 3-class-based classification models are 98.55% and 96.83%, respectively. The results also revealed that our models achieved competitive performance compared with the existing heavyweight models while significantly reducing cost and memory requirements for computing resources. With these findings, we can indicate that our models are helpful to clinicians in making insightful diagnoses of COVID-19 and are potentially easily deployable on devices with limited computational power and resources.
Keywords: COVID-19 detection, X-ray imaging, Convolution neural network (CNN), Lightweight deep learning techniques
1. Introduction
Coronavirus disease 2019, also known as COVID-19, is an infectious disease caused by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). It is highly contagious and can be passed on through airborne and respiratory droplets after coming into contact with infected patients who cough, sneeze, or even converse (Alshazly et al., 2021, Hasan et al., 2022). It was first detected in Wuhan, Hubei Province, China, on December 31, 2019, and has since quickly spread across the globe (Aakansha Gupta & Katarya, 2021a). Patients with COVID-19 infection most frequently develop symptoms such as fatigue, dry cough, fever, headache, loss of taste and smell, and respiratory illnesses like shortness of breath (M. Khan et al., 2021). So far, more than 680 million individuals worldwide have contracted this virus, and about 6.8 million have died as a result (coronavirus worldometer, 2022). Despite the virus not causing a high death toll, its impact on the world's health and well-being has been catastrophic. Therefore, many governments have strived to increase the diagnostic capacities of their hospitals so that those infected can be isolated early and receive immediate care to curb the spread of the virus (Abdani et al., 2020).
Currently, the standard screening test for COVID-19 is reverse transcription polymerase chain reaction (Minaee et al., 2020). However, this test kit is time-consuming (the results typically appear within hours to days). Besides, it has a low sensitivity rate of about 63.0% and is not widely available, particularly in developing countries (Haghanifar et al., 2022; W. Wang et al., 2020). Thus, it is necessary to consider alternative diagnostic procedures to identify patients with COVID-19 so that many lives can be saved. Fortunately, chest radiography imaging techniques have been found to be effective in this regard. This is because the virus primarily infects the lungs, causing abnormalities in chest radiography images that aid in distinguishing healthy individuals from those infected with COVID-19 (Chung et al., 2020).
Computed tomography (CT) scans and chest X-rays are two imaging modalities currently used in clinical practice to screen and diagnose COVID-19 (Hertel and Benlamri, 2022, Thakur et al., 2022). Compared with CT scans, chest X-rays are more widely used in healthcare institutions due to their low radiation dose, ease of use, low cost, and widespread accessibility. However, manually analyzing a large volume of chest X-ray images to distinguish COVID-19 subjects from others is time-consuming and challenging (Ieracitano et al., 2022, Narin et al., 2021). Moreover, the accurate diagnosis of COVID-19 infection is highly dependent on the expertise of radiologists, which poses difficulties for some due to its recent emergence and similarity to other lung diseases, such as pneumonia (Luz et al., 2022). Thus, such issues necessitate the development of an automated decision support system to improve the diagnosis's efficiency, accuracy, and speed, which can be achieved using machine learning and statistical algorithms (Jalali et al., 2022).
The significance of these algorithms in predicting and controlling the spread of COVID-19 is reflected in several studies. Some of these studies include predicting the potential impact of the virus, such as the possibility of a third wave in India (Aakansha Gupta & Katarya, 2023), analyzing Twitter sentiments related to COVID-19 using machine learning (Katarya et al., 2022), predicting COVID-19 cases based on LSTM (Long Short-Term Memory) and SIR (Susceptible-Infected-Removed) models using social media (Aakansha Gupta & Katarya, 2022), conducting a parameter-based literature survey of COVID-19 mortality dynamics using machine learning techniques (Sewariya & Katarya, 2021), reviewing various mathematical and deep learning-based forecasting methods for COVID-19 pandemic (Katarya et al., 2021), and proposing a novel LDA-based framework to forecast COVID-19 trends (Aakansha Gupta & Katarya, 2021b).
Among the various machine learning algorithms proposed in the literature, those based on deep learning (DL) have been shown to achieve impressive results in many COVID-19 prediction and control applications. DL is a subset of machine learning that mainly focuses on algorithms based on artificial neural networks, which aim to automatically extract and classify features from input data using multiple processing layers. In recent years, it has become a well-established discipline in the application of artificial intelligence to mine, analyze, and detect patterns from data (Apostolopoulos et al., 2020). Therefore, many DL algorithms, mostly convolutional neural networks (CNNs), have recently been proposed for faster and more accurate detection of COVID-19 infection through chest X-ray images. For instance, (Apostolopoulos & Mpesiana, 2020) made use of several state-of-the-art pre-trained DL models, such as VGG19, MobileNet-v2, Inception, Xception, and Inception ResNet v2, for the automatic diagnosis of COVID-19 from chest X-ray images. They tweaked these pre-trained models to distinguish COVID-19, bacterial pneumonia, and normal subjects from each other. Experimental results revealed that among the five models investigated in their study, VGG19 produces the highest accuracy rate, which is 93.48%.
(Chaudhary & Pachori, 2021) developed a novel feature extraction approach for COVID-19 detection, utilizing a Fourier-Bessel series expansion-based decomposition (FBSED) method in conjunction with a set of pre-trained CNN models. The FBSED method was employed to decompose chest X-ray and CT scan images into sub-band images (SBIs). From each SBI, deep features were extracted using a variety of well-known models, including EfficientNet, ResNet-50, Inception-ResNet-v2, AlexNet, and NASNet. These deep features were then ensemble using fusion operations and fed to a diverse set of classifiers, including J48, Naive Bayes, Softmax, AdaBoost, and Random Forest (RF) classifier. Experimental results demonstrated the effectiveness of this methodology, achieving an accuracy of 97.6%, a sensitivity of 97.0%, a specificity of 96.5%, and an F1-score of 97.0%.
(Anunay Gupta et al., 2021) proposed a computer-aided diagnostic model for the efficient detection of COVID-19 subjects from X-ray images. They utilized a set of fine-tuned pre-trained DL models, such as ResNet-101, Inception-v3, Xception, MobileNetv2, and NASNet, to extract salient features from the inputted images. Then, they combined these models using a particular integrated stacking technique to form a new model called InstaCovNet-19. Experimental results showed that Gupta's model achieved an accuracy of 99.08% in three classes (Covid-19, Pneumonia, and Normal) and 99.53% in two classes (Covid and Non-Covid).
(George et al., 2023) developed a new VGG-inspired deep CNN model for identifying COVID-19 patients using chest X-ray images. They used a combination of Contrast Limited Adaptive Histogram Equalization (CLAHE) and a homomorphic transformation filter to process and extract features from the X-ray images. Then, they fed the processed images into their newly proposed model, which was designed to handle grayscale images and perform both binary and multiclass classification. Experimental results on two public datasets showed that their new model achieved 97.41% accuracy for multi-class classification and 98.06% accuracy for binary classification.
(Heidari et al., 2020) studied the impact of two preprocessing algorithms on the performance of a pre-trained DL model, namely VGG16, after being fine-tuned for COVID-19 diagnosis using chest X-ray images. They applied a histogram equalization algorithm and a bilateral low-pass filter on the original grayscale images to generate pseudo-color images and then fed them into the three channels of the fine-tuned VGG16 model. Experimental results on the X-ray dataset of 3-class based revealed that image preprocessing algorithms help improve deep learning model performance.
(Sahin, 2022) developed a new CNN model and fine-tuned two pre-trained DL models, namely MobileNetv2 and ResNet50, to automatically detect COVID-19 patients using chest X-ray images. The developed models were designed for two-class classification (COVID-19 and Normal). Results from the experiments demonstrated that their newly developed CNN model outperforms most existing DL algorithms reported in the literature.
(Ukwandu et al., 2022) proposed three lightweight CNN models through fine-tuning MobileNetV2 architecture to detect COVID-19 infection using chest X-ray images. The first model was designed for 3-class categorization (COVID-19, Normal, Viral Pneumonia infection). Whereas the second and third ones were designed for 2-class categorization (COVID-19 and Normal, COVID-19 and Viral Pneumonia). Results indicated that their proposed models yield competitive performance to existing heavyweight models but deliver significantly greater deployment efficiency.
(Zebin & Rezvy, 2021) utilized the concept of transfer learning to tweak three pre-trained DL models for the classification of COVID-19, pneumonia, and normal class from X-ray images. Their study was limited to the following models: ResNet50, VGG-16, and EfficientNetB0. Before the training stage, they augmented the minority class using a generative adversarial framework. Results from their experiments showed that the EfficientNetB0 model performs better than the VGG16, and ResNet50 models, achieving an accuracy rate of 96.8%.
Despite the promising results of CNN models reported in previous studies, several limitations have yet to be addressed. One major limitation is that most existing models make use of pre-trained models trained on RGB image datasets, which are not appropriate for single-channel X-ray images. This can be addressed by either replicating the one-channel image data to three channels, which increases the number of operations and introduces redundancy or by altering the input layer of the pre-trained model to accept one-channel image data, which weakens the use of pre-trained weights in a transfer learning approach. Another limitation is the computational expense of these models, which have millions of parameters that need to be fine-tuned, making them impractical for devices with limited resources. Lastly, a significant challenge when using DL models is collecting an adequate number of samples with appropriate annotations for effective training, and most of the earlier models in the literature were trained and validated on fairly small datasets with a limited number of X-ray images.
Motivated by the limitations of the studies mentioned above, we in this research developed two new lightweight CNN-based diagnostic models for the automated and early detection of COVID-19 from chest X-ray images. The first model was designed for binary classification, specifically to distinguish between COVID-19 and normal classes. On the other hand, the second model was designed for multiclass classification, such as distinguishing between COVID-19, viral pneumonia, and normal classes. The purpose of using a lightweight deep CNN in our models was to reduce the number of learnable parameters and computations required. Additionally, using a lightweight model can also result in faster training and inference times, which is important in real-world applications where time constraints may be an issue. Despite their simplicity, the proposed models provided promising results and required significantly fewer tuning parameters when applied to a relatively large dataset.
The main contributions of this study can be summarized as follows:
-
•
Two lightweight CNN models were developed for the early detection of COVID-19 subjects from chest X-ray images. The first is for detecting either COVID-19 or Normal subjects, whereas the second is for detecting either COVID-19, Viral Pneumonia, or Normal subjects.
-
•
The proposed models were tested on a fairly large dataset, and the results obtained using various classification metrics were encouraging and promising.
-
•
To the authors' knowledge, the proposed models are the first to include a small number of learnable parameters while still achieving state-of-the-art performance. Hence, such models are advantageous for systems with low computational power, such as cell phones and medical diagnostic devices.
-
•
Compared with the up-to-date deep learning models reported in the literature, our models greatly reduce the computational power and memory resources needed during the classification process. In addition, they demonstrated better performance in terms of accuracy rate than most of the existing models.
The remaining sections of this paper are structured as follows. Section 2 describes the dataset used in this research and the proposed models for COVID-19 detection. It also presents the performance measures used to evaluate our models. Section 3 reports and discusses the results of our experiments, and it also provides a detailed performance analysis of our models against other well-established ones found in the literature. Finally, in Section 4, we conclude the paper.
2. Materials and methods
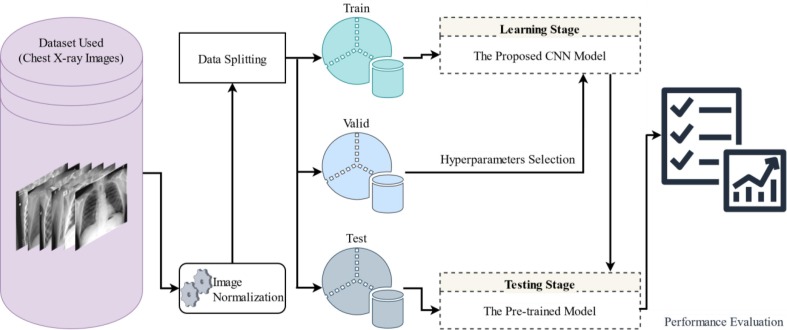
Fig. 1 illustrates the methodology followed by this study for distinguishing COVID-19 patients from other subjects using chest X-ray images. The following subsections provide detailed descriptions of each step shown in Fig. 1.
Fig. 1.
Schematic diagram of the proposed methodology.
2.1. Dataset description
An open-access dataset, shared publicly by the Kaggle platform (https://www.kaggle.com/datasets/tawsifurrahman/covid19-radiography-database) (COVID-19 radiography database, 2022), was used to perform our experiments. This dataset was developed by a team of researchers from Dhaka University in Bangladesh, and the University of Qatar in Doha, Qatar, in conjunction with their collaborators from Malaysia and Pakistan, under the supervision of medical professionals. The aim of their endeavor was to provide a comprehensive and accurate dataset that can be utilized for research and analysis in the field.
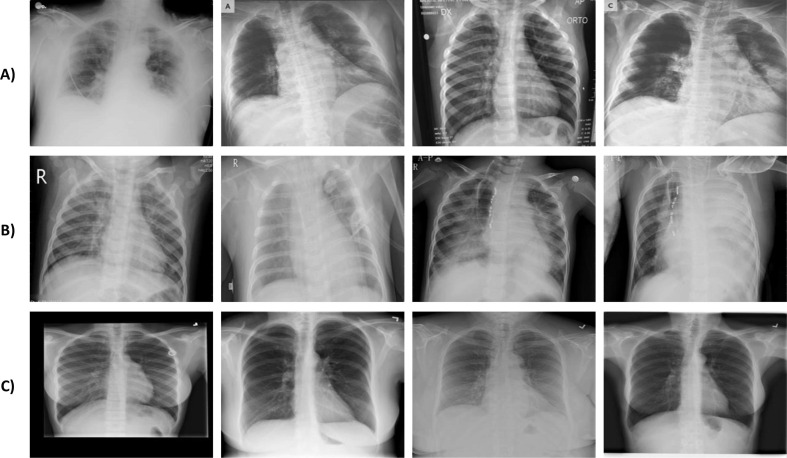
The dataset includes chest X-ray images of COVID-19, Viral Pneumonia, and Normal subjects. At the time of the study, the categories mentioned earlier contained 3,616, 1,345, and 10,192 samples, respectively. Images of the X-rays have a standard size of 299 × 299 pixels and were taken from different angles and positions, as shown in Fig. 2 . In this study, we resized the images to 112 × 112 because this size produced similar results as other possible larger sizes. Thereby, it helped speed up both the training and testing processes.
Fig. 2.
Some samples of chest X-ray images: A) COVID-19, B) Viral Pneumonia, and C) Normal cases.
2.2. Image normalization and data splitting
Normalizing image data before passing them on to deep learning models has become an essential step toward a faster learning process and numerical stability. For instance, when image data are normalized, the learning algorithm is more likely to converge faster (Nayak et al., 2021). Hence, in this study, the input image data () were normalized to range between [-1, 1] using Eq. (1).
| (1) |
The normalized images were then randomly partitioned into three independent sets: training, validation, and testing, as shown in Table 1 . This is to validate the efficacy of the proposed models for COVID-19 detection. The data splitting strategy was performed by initially dividing the dataset described in Section 2.1 into training and testing sets with a ratio of 80%:20%, respectively. Subsequently, the training dataset was divided into training and validation sets with the same ratio mentioned earlier.
Table 1.
The number of instances used as the training, validation and testing sets.
| Class Label | Training | Validation | Testing |
|---|---|---|---|
| Covid-19 | 2314 | 579 | 723 |
| Normal | 6522 | 1631 | 2039 |
| Pneumonia | 861 | 215 | 269 |
2.3. Proposed lightweight CNN models
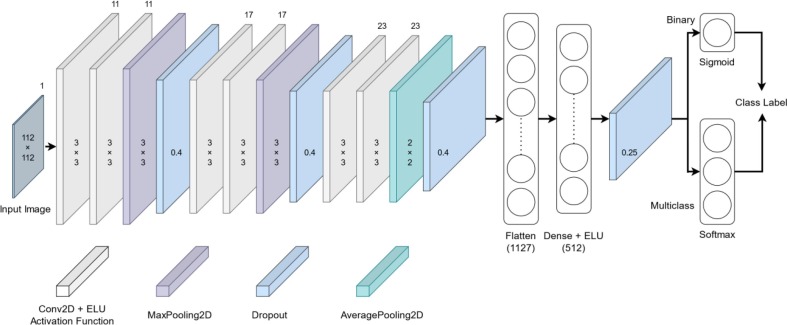
The architecture of the proposed lightweight CNN models for the automatic classification of COVID-19 patients is depicted in Fig. 3 . These models are basically two models, sharing the same hidden layer structure (feature extraction layers) except for the output layer (classification layer). The first model is 2-class based, i.e., COVID-19 and normal. Whereas the second one is 3-class based, i.e., COVID-19, viral pneumonia, and normal. Both models are primarily designed to detect COVID-19 patients from chest X-ray images. The purpose of developing such models is to reduce the computational cost and memory requirements while still achieving state-of-the-art performance. This can be particularly useful in applications where the model needs to be deployed on resource-constrained devices, such as mobile phones or embedded systems. Additionally, lightweight models are faster to train and predict, making them more convenient to use in real-world applications. In the following subsections, we first provide an overview of CNN model. Then, we present a detailed description of the architecture, training and testing phases of the aforementioned proposed models.
Fig. 3.
Structure of the proposed lightweight CNN models for detecting COVID-19 patients.
2.3.1. CNN: An overview
CNN is a particular type of artificial neural network designed to process data similar to that seen by the human visual system. It has recently been used in a variety of visual recognition tasks, such as image classification (Hussain et al., 2021) and object detection (Dhillon & Verma, 2020), and has proven to produce impressive results. A typical CNN model consists of three main layers: convolution layers, pooling layers, and fully connected layers.
A. Convolutional layer
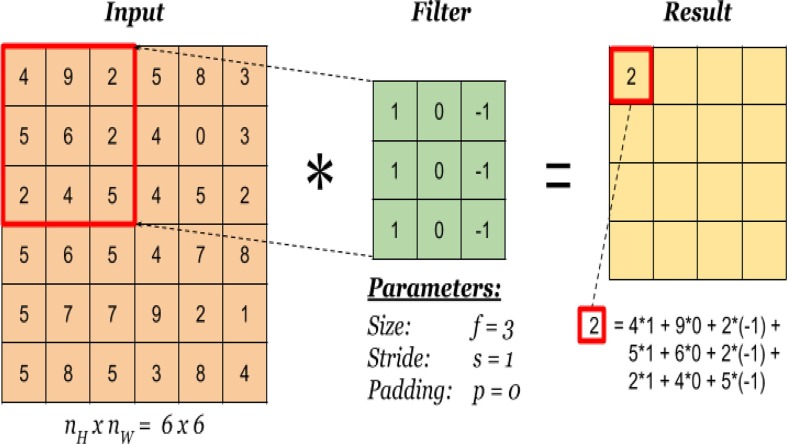
It is one of the core layers of CNN model, which constructs a certain number of filters (kernels) with a fixed size (such as 3 × 3, 5 × 5, or 7 × 7) that are convolved over input images to perform feature extraction. The convolution operation is performed by first dividing an input image into blocks (spaced according to the stride set), each being of the same size as the filter to be applied to. Subsequently, an element-wise multiplication between each block (left to right, top to bottom) and the given filter is performed and then summed to produce a single output value (Fig. 4 ). The output values from the previous step are placed in the corresponding position of the output matrix, named a feature map. By repeating the above process with all the given filters, a number of feature maps are formed, representing different characteristics of the input image (Yamashita et al., 2018).
Fig. 4.
Illustration of the convolution operation on an image of 6 × 6 pixels (Convolution operation, 2022).
An interesting feature of the convolution operation is weight sharing: the same filters are applied to all different spatial locations of the image. With this property, fewer weights are required to learn, decreasing the time and cost of training the model. To make the output of the convolution operation nonlinear, an activation function, such as ELU or ReLU, is applied to it. This activation function makes the model learn salient features that best describe the data of interest, thereby boosting the classification performance.
B. Pooling layer
The feature maps obtained after the convolution process contain too many features that are susceptible to causing an overfitting issue. To overcome this issue, the concept of pooling (also known as down-sampling) was introduced. With pooling, the feature maps are reduced in size, leading to drastic lower computational costs for the subsequent layers. Aside from that, the pooling layer reduces the total number of parameters that the network has to learn (Li et al., 2021).
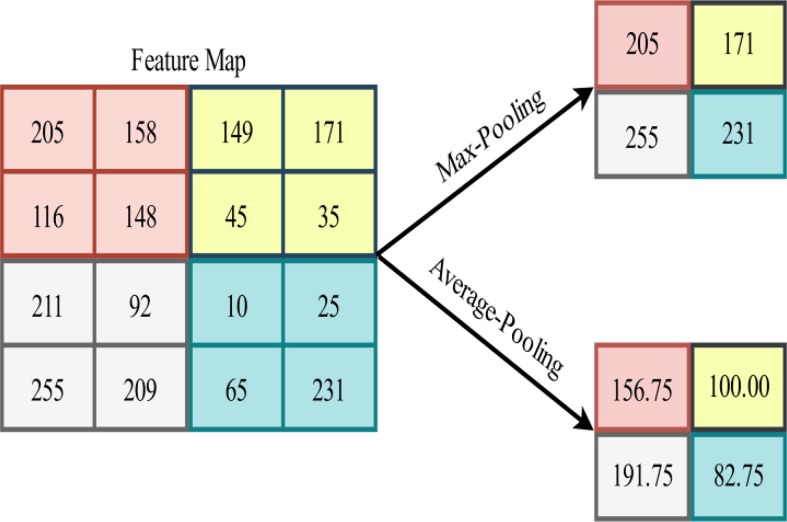
The most widely used forms of pooling are Max-Pooling and Average-Pooling. The former partitions the feature maps into patches and then extracts the maximum value of each patch to obtain down-sampled feature maps. The latter, on the other hand, computes the mean value for patches of the feature maps. Fig. 5 provides a pictorial representation of the aforementioned pooling operations.
Fig. 5.
Illustration of Max-Pooling and Average-Pooling operations applied to an arbitrary feature map.
C. Fully connected layer
The output of the feature maps obtained by the last convolutional or pooling layer is transformed into a 1-D feature vector using the flatten layer. The main reason for this is that the classification layer (i.e., sigmoid or softmax) requires its input to be in that form. This layer is represented by a fully connected layer (also known as a dense layer), which connects each of its internal nodes (or neurons) to each incoming input node by a learnable weight. The number of nodes in the classification layer depends on the particular task under consideration. For instance, if the data to be studied consists of two classes, the number of nodes is two with a softmax activation function, however the number changes to one with a sigmoid activation function (Aggarwal et al., 2022). It should be noted that sigmoid activation is not used in the classification layer when the data consists of more than two classes.
Usually, a few fully connected layers with a predefined number of nodes are placed between the flatten layer and the classification layer to serve as feature extractors. These layers are also followed by a nonlinear activation function, e.g., ELU or ReLU, which helps the network learn nonlinear combinations of high-level features.
2.3.2. Architecture of the proposed models
Table 2 presents a summary demonstrating the architecture of the developed models for COVID-19 detection. As mentioned previously, the feature extraction part of both proposed models is identical, which consists of four hierarchical blocks. The first block consists of two consecutive convolutional layers, each containing 11 filters of 3x3 size, a stride of 1, and padding='same', followed by a Max-Pooling layer with a pool of 3x3 size, a stride of 3, and padding='same'. The second block consists of the same layers as the first block except for the number of filters used, which is 17. The third block consists of two consecutive convolutional layers, each containing 23 filters of 3x3 size, a stride of 1, and padding='same', followed by an Average-Pooling layer with a pool of 2x2 size, a stride of 2, and padding='same'. The last block consists of a fully connected layer containing 512 neurons, which receives the flattened version of its previous block output as an input.
Table 2.
Architecture of the proposed CNN models for COVID-19 prediction.
| Layer | Filter | Padding | Activation | Kernel Initializer | Output Shape | Param # |
|---|---|---|---|---|---|---|
| Input | – | – | – | – | (112, 112, 1) | 0 |
| Conv2D (3 × 3) | 11 | Same | ELU | HeNormal | (112, 112, 11) | 110 |
| Conv2D (3 × 3) | 11 | Same | ELU | HeNormal | (112, 112, 11) | 1100 |
| MaxPooling2D (3 × 3) | – | Same | – | – | (38, 38, 11) | 0 |
| Dropout (0.4) | – | – | – | – | (38, 38, 11) | 0 |
| Conv2D (3 × 3) | 17 | Same | ELU | HeNormal | (38, 38, 17) | 1700 |
| Conv2D (3 × 3) | 17 | Same | ELU | HeNormal | (38, 38, 17) | 2618 |
| MaxPooling2D (3 × 3) | – | Same | – | – | (13, 13, 17) | 0 |
| Dropout (0.4) | – | – | – | – | (13, 13, 17) | 0 |
| Conv2D (3 × 3) | 23 | Same | ELU | HeNormal | (13, 13, 23) | 3542 |
| Conv2D (3 × 3) | 23 | Same | ELU | HeNormal | (13, 13, 23) | 4784 |
| AveragePooling2D (2 × 2) | – | Same | – | – | (7, 7, 23) | 0 |
| Dropout (0.4) | – | – | – | – | (7, 7, 23) | 0 |
| Flatten | – | – | – | – | (1127) | 0 |
| Dense | – | – | ELU | HeNormal | (5 1 2) | 577,536 |
| Dropout (0.25) | – | – | – | – | (5 1 2) | 0 |
| Output of Binary Classification | ||||||
| Dense | – | – | Sigmoid | HeNormal | (1) | 513 |
| Output of Multiclass Classification | ||||||
| Dense | – | – | Softmax | HeNormal | (3) | 1539 |
After each of the above blocks, a dropout layer was added with a drop rate reported in Table 2. During training time, this layer randomly sets input units to zero with a specific drop rate at each step, which helps prevent complex co-adaptation to the training data. Furthermore, the output of the convolutional layers and the fully connected layer was activated using the ELU function defined in Eq. (2).
| (2) |
where is the input tensor to be activated, and is a constant value help control the value at which an ELU saturates for the input tensor's negative values. In this study, we set to 1 experimentally.
Lastly, the high-level features obtained from the above blocks were fed to each of the two proposed classifiers for classification. The first classifier (i.e., 2-class based) is a fully connected layer with a single neuron followed by the sigmoid activation function. This function is expressed mathematically as in Eq. (3), which produces an output that falls in the range of (0, 1). Eq. (4) was used as the decision-making of this classifier.
| (3) |
| (4) |
As for the second classifier (i.e., 3-class based), we used a fully connected layer with three neurons followed by the softmax activation function. This function is expressed mathematically as in Eq. (5), which generates a probability score for each class, with all probabilities summed to 1. During prediction, this classifier predicts the class with the highest probability.
| (5) |
where is 3, indicating the number of classes, and is the input tensor containing the probabilities of the classes.
While building the proposed models, the most challenging part was combining all the aforementioned layers and activation functions to obtain excellent performance in terms of accuracy and other evaluation metrics. In particular, we focused on building a cost-effective architecture to reduce the computational complexity of the network while maintaining a high level of accuracy. This was achieved by using smaller filters, fewer layers, and lower-dimensional representations, as evidenced by Table 2, which shows a reasonable number of learnable parameters of 591,903 and 592,929 for the proposed binary and multiclass classification models, respectively.
2.3.3. Model training and testing
To train and test the two proposed models, we initially normalized the chest X-ray images and then divided them into three sets, namely training, validation, and testing, as described in Section 2.2. We subsequently trained both models on the training set and determined the model's best hyperparameters using the validation set. Finally, we evaluated both models on the testing (unseen) set using various performance metrics described in the following subsection. It should be noted that the 2-class-based model was trained and tested only on the COVID-19 and Normal classes reported in Table 1.
During training, the binary and categorical cross-entropy losses were used for the binary and multiclass models. These loss functions measure the deviation of the values predicted by the model from the true values, which enables the model to improve over successive epochs by reducing the error in the predictions using a particular optimization algorithm. The above losses are expressed mathematically as in Eqs. (6), (7).
| (6) |
| (7) |
where and are the binary and categorical cross-entropy losses, respectively, refers to the number of samples (chest X-ray images) used, and represent the true and predicted values of the samples, and is the number of classes.
In this study, we used an Adam optimizer to optimize the learnable parameters of both proposed models, which is a variant of stochastic gradient descent. Furthermore, we employed the class weight method (Heidari et al., 2020) to handle the imbalanced dataset shown in Table 1, which was applied to the training data during the learning phase to minimize the potential consequences on the model’s performance. This method assigns higher weights to the minority classes than the majority ones to compensate for their underrepresentation during training. Thus, the loss calculation becomes a weighted average, in which each sample is given a weight corresponding to its associated class. Eq.(8) was used to compute the weight of each class. Details about other hyperparameters used during model training can be found in Table 3 . Furthermore, a summary of the steps needed for implementing the proposed models is provided in Algorithm 1.
| (8) |
where is the weight of class , is the total number of training samples, is the number of classes, and is the number of samples belonging to class .
| Algorithm 1: Pseudocode of the proposed COVID-19 diagnostic models |
|---|
Input: Training, validation, and test sample sets: ,andOutput: Predicted Binary class label vector:Predicted Multi-class label vector:
|
Table 3.
Hyperparameters used during training.
| Hyperparameter | Value |
|---|---|
| Optimizer | ‘adam’ |
| Learning_rate | 0.001 |
| Beta_1 | 0.9 |
| Beta_2 | 0.999 |
| Class_weight | {0: 1.9092, 1: 0.6774} for binary model |
| {0: 1.3969, 1: 0.4956, 2: 3.7542} for multiclass model | |
| Epoch | 100 |
| Batch size | 32 |
2.3.4. Evaluation metrics
Detecting COVID-19 patients from chest X-ray images is, in essence, a classification problem. Therefore, we selected some commonly used classification metrics derived from the confusion matrix to measure the effectiveness of the proposed models. Accuracy (Alghamdi et al., 2021), sensitivity (Bhattacharyya et al., 2022), specificity (Akter et al., 2021), and Matthews correlation coefficient (MCC) (Ali et al., 2021) are the metrics used in this study, which are expressed mathematically as in Eqs. (9)-(12), respectively.
| (9) |
| (10) |
| (11) |
| (12) |
where (Correct Samples) represents the number of samples being correctly predicted, (Total Samples) represents the total number of samples to be predicted, (True Patient) and (True Normal) refer to the number of subjects that were correctly predicted as COVID-19 and normal. (False Patient) and (False Normal) indicate the number of subjects that were incorrectly predicted as COVID-19 and normal.
Besides the above metrics, we used another metric called the area under the curve (AUC) of receiver operating characteristic (ROC). This metric shows the sensitivity of a particular classifier to the threshold value used during the classification process.
3. Results and discussion
In this section, we comprehensively evaluated the proposed lightweight CNN models for COVID-19 detection using widely adopted performance measures. The proposed models were implemented using the Python programming language installed on a computer equipped with Intel(R) Core(TM) i7-8550U CPU @1.80 GHz 1.99 GHz, 12 GB of RAM, and Windows 11 as an operating system. In the following subsections, we first report and discuss the results obtained by the proposed models. Then, we present a comparative analysis between our proposed models and the latest existing ones.
3.1. Accuracy and loss vs. Epoch
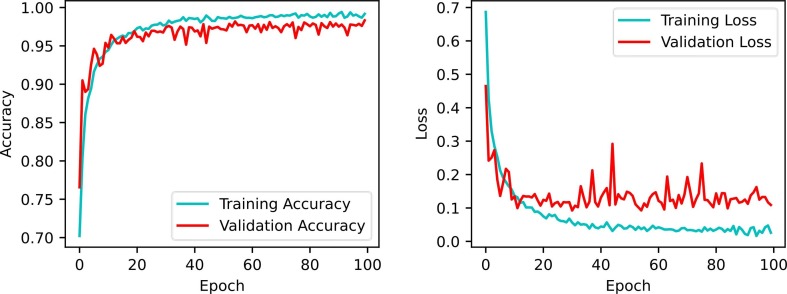
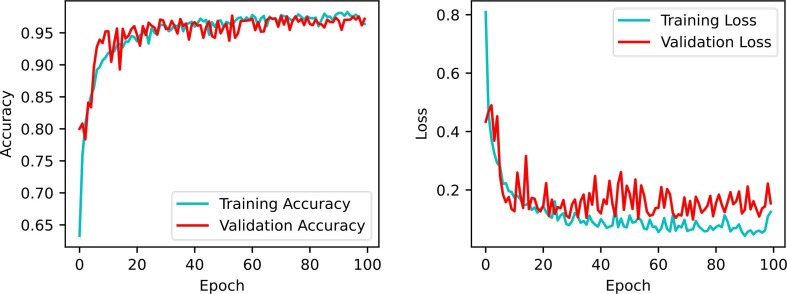
During the training stage of a given classifier algorithm, accuracy and loss are the most common metrics chosen to be displayed over successive epochs to control overfitting and perceive the predictions' status. These metrics are assumed to be inversely proportional, i.e., with low loss values, high accuracy values are expected. Results of the aforementioned metrics on the training and validation sets are illustrated in Fig. 6, Fig. 7 . The trend curves of these figures indicate that as the number of epochs increases, the accuracy and loss of the validation set initially vary significantly with notable fluctuations but then gradually improve to yield higher accuracy and lower loss with small fluctuations. The trend curves also show that validation accuracy tends to be close to training accuracy over multiple epochs, indicating that the proposed models do not show signs of significant underfitting or overfitting.
Fig. 6.
Performance of accuracy and loss during model training and validation for binary classification.
Fig. 7.
Performance of accuracy and loss during model training and validation for multiclass classification.
3.2. Confusion matrix analysis
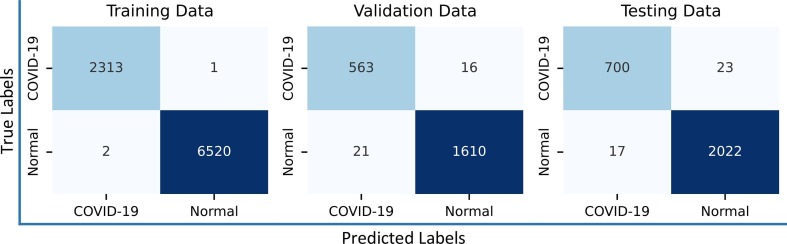
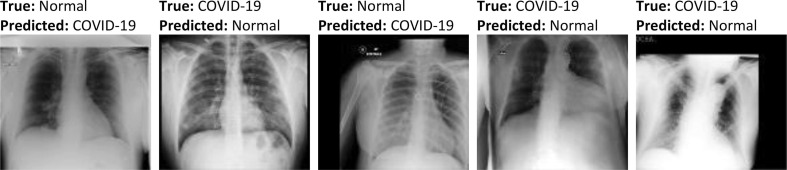
This matrix provides a summary containing the prediction results of a given classification algorithm. Its name derives from the fact that it makes it easy to observe where the model gets confused. That is, mislabeling some samples by another class label. Fig. 8 shows results of the confusion matrices obtained by the proposed 2-class based model. As can be seen from Fig. 8, the binary model misclassified 1, 16, and 23 COVID-19 samples from the training, validation, and testing sets as belonging to the Normal class. The error rates of these COVID-19 samples are 0.04%, 2.76%, and 3.18%. Similarly, 2, 21, and 17 Normal samples were misclassified as belonging to the COVID-19 class, resulting in an error rate of 0.03%, 1.29%, and 0.83% in samples of the Normal class. Some samples that were misclassified by this diagnostic model is shown in Fig. 9 . A possible reason for the misclassification could be the similar imaging characteristics between COVID-19 and normal samples, as well as the misalignment and non-frontal view of some of these samples. Considering the low misclassification rates in the above classes, one can tell that the proposed model (2-class-based) achieves excellent results, mainly due to its ability to generate features with high interclass discrimination.
Fig. 8.
Confusion matrices obtained by the proposed binary classification model.
Fig. 9.
Some samples of misclassified chest X-ray images obtained by the proposed binary classification model.
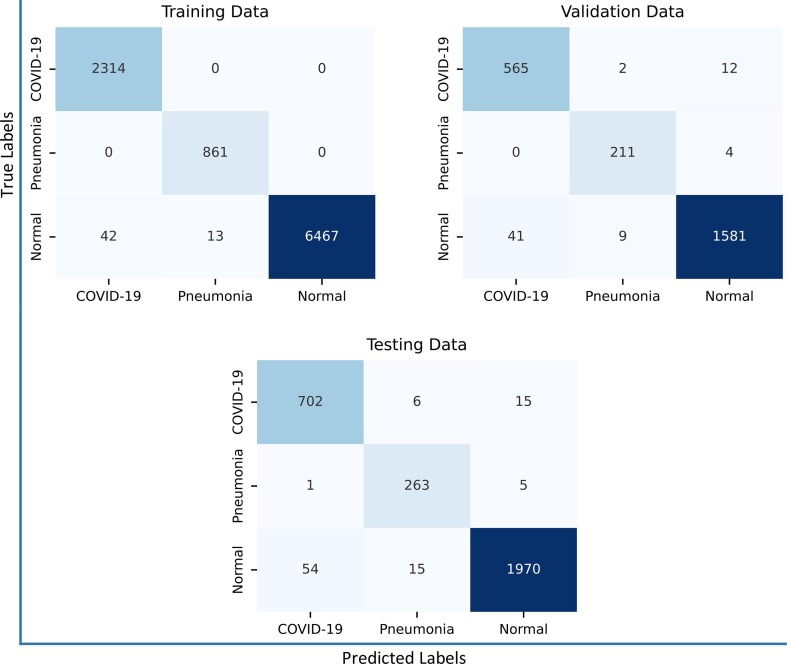
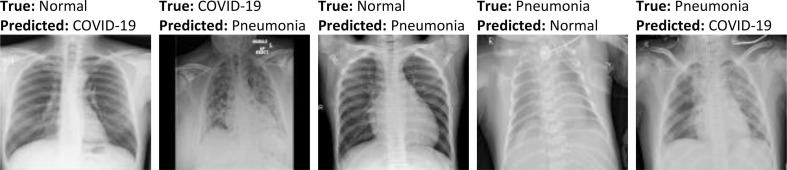
As for the proposed 3-class-based model, the confusion matrices generated by it are shown in Fig. 10 . From this figure, it is evident that the samples of Normal and COVID-19 classes are more likely to be mislabeled than samples of Viral Pneumonia class. This is mainly due to the high similarity between the features of some samples of these categories (Fig. 11 ), making differentiation extremely difficult. However, the overall performance remains satisfactory. For instance, from the confusion matrix of the testing set, it is quite clear that only 0.83% and 2.07% of COVID-19 samples were misclassified as Viral Pneumonia and Normal, respectively. Likewise, 0.74% and 2.65% of Normal samples were mislabeled as Viral Pneumonia and COVID-19, respectively. With these results, one can conclude that the proposed model (3-class-based) yields a very low error rate in each of the aforementioned categories, demonstrating its remarkable ability to learn features.
Fig. 10.
Confusion matrices obtained by the proposed multiclass classification model.
Fig. 11.
Some samples of misclassified chest X-ray images obtained by the proposed multiclass classification model.
3.3. Overall performance of the proposed models
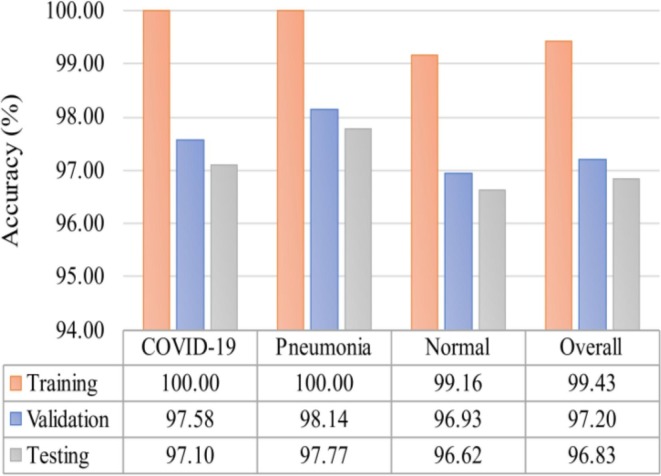
We obtained further results using some widely reported evaluation metrics to examine the overall performance of the proposed 2-class and 3-class classification models. These results can be found in Table 4 and Fig. 12 . From Table 4, it can be seen that the overall accuracy is 99.97% (training), 98.33% (validation), and 98.55% (testing) for the 2-class-based model. As for the 3-class-based model, the overall accuracy is presented in Fig. 12, which is 99.43% (training), 97.20% (validation), and 96.83% (testing). The accuracy results suggest the feature extraction part of both proposed models is highly discriminatory as the results obtained are promising. Furthermore, when both models are compared, it is evident that the 2-class classification model outperforms the 3-class classification model. This is expected because the multiclass labels are more difficult to distinguish than the binary class labels.
Table 4.
Performance analysis of the proposed binary classification model under different evaluation metrics.
| Metrics | Training | Validation | Testing |
|---|---|---|---|
| Accuracy (%) | 99.97 | 98.33 | 98.55 |
| Sensitivity (%) | 99.96 | 97.24 | 96.82 |
| Specificity (%) | 99.97 | 98.71 | 99.17 |
| MCC | 0.999 | 0.957 | 0.962 |
Fig. 12.
Accuracy performance of each class along with the overall accuracy result of the proposed multi-class classification model.
Since the dataset used in this study is imbalanced, considering accuracy as the sole performance metric is misleading. For instance, the class with the most samples is more likely to suppress the other classes, affecting the model's overall performance. Therefore, we report other popular metrics used for this purpose. It can be noticed from Table 4 that the sensitivity and specificity of the unseen (testing) data obtained by the proposed 2-class classification model are 96.82% and 99.17%, respectively. Such results indicate the remarkable performance of the proposed model because there is no significant bias toward the Normal class over the COVID-19 class. That is, the specificity of the Normal class does not differ much from the sensitivity of the COVID-19 class. Furthermore, the MCC obtained by the binary model is 0.962, which again confirms that there is no bias due to its closeness to the optimal value, i.e., 1.
As for the proposed 3-class classification, we report the accuracy of each class to determine its overall performance. The accuracy result of each class is listed in Fig. 12. Taking the testing set as an example, we can observe that the accuracy of the three classes is very close to one another, i.e., 97.10% for COVID-19, 97.77% for Pneumonia, and 96.62% for Normal. Despite the slight differences in their performance, the results of the three categories are very satisfactory. These results clearly demonstrate that the proposed model for multiclass classification does not favor any class over others.
Even though both proposed models were trained on imbalanced training data, the above results show no evidence of bias. We believe this is mainly due to applying the class weight technique to the training data. With this technique, different weights are assigned to different classes, with the highest weight being assigned to the class with the fewest samples. This is to help the loss function in making the minority class's cost higher than the majority class's cost when they are incorrectly predicted, thereby forcing the learning algorithm to be unbiased.
3.4. ROC curve analysis
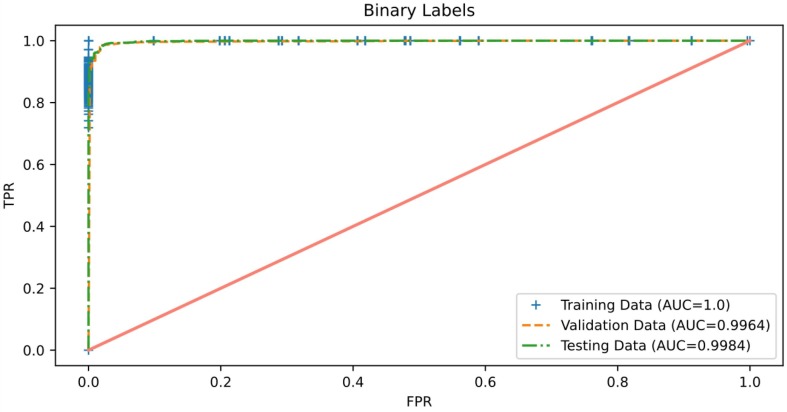
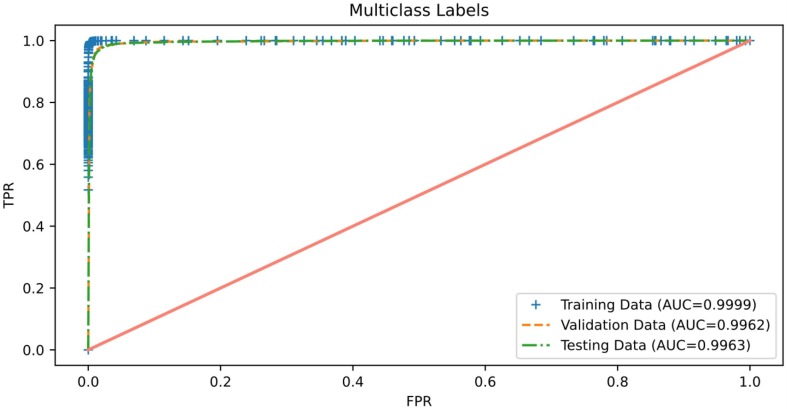
The ROC curve is a graph that demonstrates the classifier's ability to distinguish between different class labels against different thresholds used during prediction. Therefore, we consider reporting this graph to illustrate the classifier's effectiveness at discriminating COVID-19 from other cases. Fig. 13, Fig. 14 report results of the AUC of ROC obtained by the proposed binary and multiclass classification models, respectively. It can be observed from these figures that both proposed models perform extremely well in terms of AUC on the three sets of chest X-ray images: training, validation, and testing, in which an AUC value of more than 0.99 was achieved for each data partitioning set of both models. These findings indicate that both proposed models are almost invariant to the thresholds used during prediction because the AUCs obtained are very close to the optimal value, i.e., 1. Thus, we can infer that the frameworks proposed for COVID-19 detection seem to have excellent deployment potential.
Fig. 13.
Roc curve of the proposed 2-class classification model.
Fig. 14.
Roc curve of the proposed 3-class classification model.
3.5. Run time complexity analysis
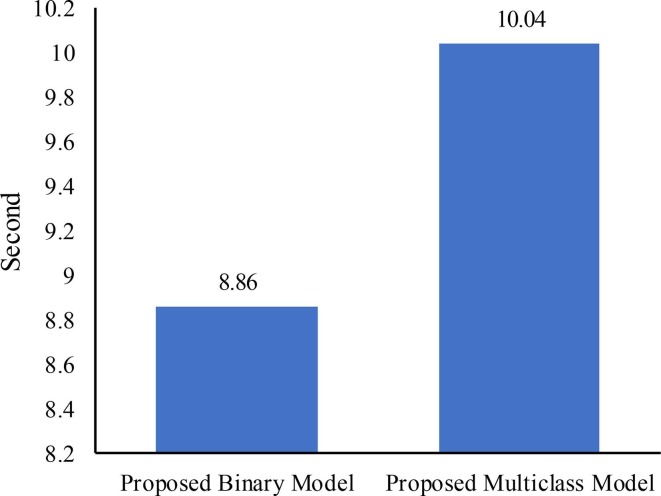
The computational complexity of the proposed models has been estimated in relation to their crucial role in DL models. The exponential growth in computational complexity is directly related to the increase in network-level growth (Fang & Wang, 2022). Determining the computational complexity usually involves leveraging the trainable parameters from the model's architecture (Ahrabi et al., 2021). Hence, in this study, the trainable parameters within the proposed model's architecture were used to determine the computational complexity. The proposed models have been shown to require a reasonable number of trainable parameters, as demonstrated by Table 2. The models require an approximate total of 590 k trainable parameters. Moreover, Fig. 15 displays the duration required by the proposed models for predicting the class label of test data. It can be observed from this figure that the proposed binary model requires 8.86 s to predict the class label of 2762 test chest X-ray images. In contrast, the proposed multiclass model takes 10.04 s to predict the class label of 3031 test chest X-ray images. These results demonstrate the efficiency of the proposed models in terms of computational complexity and real-time adoption.
Fig. 15.
The time needed by the proposed models to predict the class labels of test data.
3.6. Comparative analysis with state-of-the-art approaches
We compared the proposed models with the recent models published in the literature for COVID-19 detection to determine how well our models fit in with them. Table 5, Table 6 present results of the comparison. It is obvious from these tables that a direct comparison of performance is not fair due to the use of different sizes of datasets in the reported studies. This issue was prevalent in many previous studies, as the availability of samples changed over time, in which samples are now becoming more abundant. For instance, it was difficult to obtain a large number of samples during the first year of the COVID-19 outbreak. Over time, however, the availability of samples began to increase and became more accessible thanks to public repositories, such as GitHub and Kaggle.
Table 5.
Performance analysis of the proposed 2-class based model with relevant existing deep learning models in terms of accuracy and trainable parameters.
| Reference | Method | Data Size | Accuracy (%) | Trainable Parameters | |
|---|---|---|---|---|---|
| COVID-19 | Normal | ||||
| (Anunay Gupta et al., 2021) | Inception-V3 | 361 | 365 | 97.00 | 24,000,000 |
| (George et al., 2023) | GrayVIC model | 2,250 | 2,250 | 98.06 | 2,684,650 |
| (Hussain et al., 2021) | CoroDet | 500 | 800 | 99.10 | 2,873,609 |
| (Jyoti et al., 2023) | ResNet50 | 2409 | 2866 | 95.67 | 23,591,810 |
| (A. I. Khan et al., 2020) | CoroNet | 284 | 310 | 99.00 | 33,969,964 |
| (Malik et al., 2022) | Vgg16 | 4630 | 1583 | 93.00 | 14,715,714 |
| (Nayak et al., 2021) | ResNet-34 | 775 | 775 | 98.33 | 21,800,000 |
| (Nayak et al., 2023) | LW-CORONet | 2358 | 8066 | 96.25 | 680,000 |
| (Sahin, 2022) | CNN_Model | 3,626 | 10,198 | 96.71 | 667,458 |
| (Ukwandu et al., 2022) | MobileNet-V2 | 1,200 | 1,341 | 99.60 | 3,538,984 |
| Proposed Model | Lightweight CNN Model | 3,616 | 10,192 | 98.55 | 591,903 |
Table 6.
Performance analysis of the proposed 3-class based model with relevant existing deep learning models in terms of accuracy and trainable parameters.
| Reference | Method | Data Size | Accuracy (%) | Trainable Parameters | ||
|---|---|---|---|---|---|---|
| COVID-19 | Normal | Pneumonia | ||||
| (Apostolopoulos & Mpesiana, 2020) | VGG19 | 224 | 504 | 714 | 93.48 | 143,667,240 |
| (Anunay Gupta et al., 2021) | Inception-V3 | 361 | 365 | 362 | 97.00 | 24,000,000 |
| (George et al., 2023) | GrayVIC model | 2,250 | 2,250 | 2,250 | 97.41 | 2,684,650 |
| (Hussain et al., 2021) | CoroDet | 500 | 800 | 800 | 94.20 | 2,874,635 |
| (Heidari et al., 2020) | VGG16 | 445 | 2,880 | 5,179 | 94.50 | 138,000,000 |
| (A. I. Khan et al., 2020) | CoroNet | 284 | 310 | 657 | 95.00 | 33,969,964 |
| (Nayak et al., 2023) | LW-CORONet | 2358 | 8066 | 5575 | 95.67 | 680,000 |
| (Ukwandu et al., 2022) | MobileNet-V2 | 1,200 | 1,341 | 1,345 | 94.50 | 3,538,984 |
| (L. Wang et al., 2020) | COVID-Net | 358 | 8,066 | 5,538 | 93.30 | 11,750,000 |
| (Zebin & Rezvy, 2021) | EfficientNetB0 | 202 | 300 | 300 | 96.80 | 5,300,000 |
| Proposed Model | Lightweight CNN Model | 3,616 | 10,192 | 1,345 | 96.83 | 592,929 |
Even though the dataset sizes are variable, it is quite clear that the proposed models outperform most of the existing models by a large margin in terms of accuracy performance, as evidenced by the results presented in Table 5, Table 6. The proposed binary model achieved an improvement in accuracy rate of 1.55%, 0.49%, 2.88%, 5.55%, 0.22%, 2.30%, and 1.84% when compared to Inception-V3, GrayVIC model, ResNet50, Vgg16, ResNet-34, LW-CORONet, and CNN_Model, respectively. Similarly, the proposed multiclass model showed an improvement in accuracy rate of 3.35%, 2.63%, 2.33%, 1.83%, 1.16%, 2.33%, 3.53%, and 0.03% when compared to VGG19, CoroDet, VGG16, CoroNet, LW-CORONet, MobileNet-V2, COVID-Net, and EfficientNetB0, respectively. However, it is noted that the models CoroDet, CoroNet, and MobileNet-V2 performed slightly better than our binary classification model, and Inception-V3 and GrayVIC models achieved a higher accuracy rate than our multiclass classification model. We believe that the aforementioned models scored better accuracy rates than ours because they used a much smaller dataset for testing their model's performance.
From Table 5, Table 6, one can also observe that the models reported in the literature require between 1.1 and 57.4 and 1.15 to 242.3 times more learnable parameters than the proposed binary and multiclass models, respectively. This indicates a remarkable achievement since our models require much less computing power and memory resources, which are necessary for building a cost-effective model.
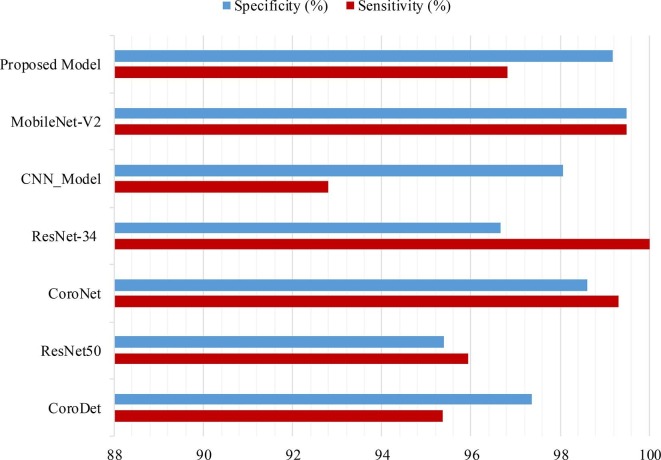
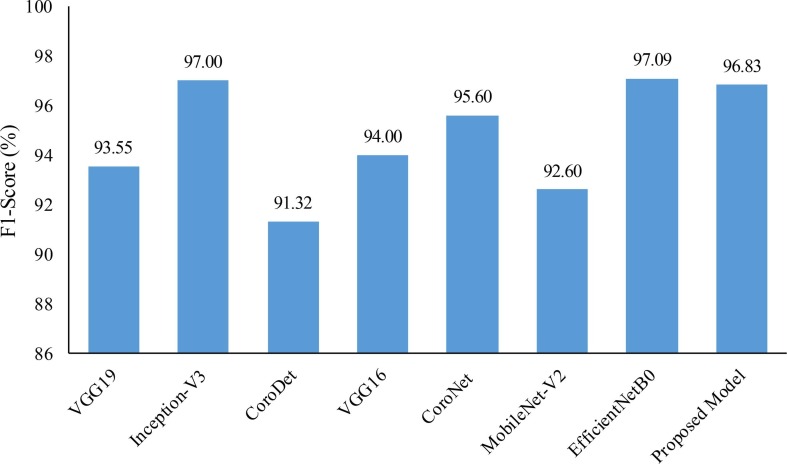
We also compared the proposed models with the recent existing models in terms of sensitivity, specificity, and f1-score metrics. Results of the comparison are presented in Fig. 16, Fig. 17 . It can be observed from these figures that both of the proposed models exhibit a significant improvement in performance when compared with many of the existing models. However, a slight edge in terms of f1-score was observed for Inception-V3 and EfficientNetB0 when compared to the proposed multiclass model. Additionally, MobileNet-V2 exhibited slightly higher values of sensitivity and specificity when compared to the proposed binary class model. We believe that the superior performance of these existing models may be attributed to their testing on small datasets. This belief is supported by the understanding that a larger dataset can lead to better generalizability of models.
Fig. 16.
Comparison of sensitivity and specificity results for binary model: Proposed One vs. Recently Published Literature.
Fig. 17.
F1-Score comparison for multiclass model: Proposed One vs. Recently Published Literature.
In summary, when comparing the proposed models with the heavyweight models, the results show that they are competitive. Furthermore, they demonstrate that our models can serve as the basis for low-cost mobile decision support applications. With these features in mind, we believe that our models can help radiologists save more lives of those infected with COVID-19 by making faster and more accurate decisions. In particular, our models are very useful for developing countries or rural areas since they have limited access to highly accredited radiologists and affordable diagnostic kits.
3.7. Limitations and future directions
Despite the satisfactory performance of the proposed lightweight CNN models, there is still scope for further improvement. One potential avenue for improvement is the application of image preprocessing algorithms to X-ray images prior to their utilization in the proposed models. However, it is important to consider that the model’s performance is also contingent upon other factors such as the quality of the data used for training and the complexity of the task at hand. Additionally, the application of certain image preprocessing algorithms may not be suitable for all types of X-ray images, and may even negatively impact the model's performance. Therefore, a comprehensive evaluation to identify image preprocessing methods that positively impact the performance of the proposed models would be a valuable area of study.
Another limitation that must be acknowledged is that neither the source of the dataset used in this study nor the source of the studies reported in the literature provides any information about the demographic characteristics of the participants. Specifically, there is no indication of the age, authenticity, or gender of the individuals included in the sample. As a result, it is challenging to determine whether the proposed models exhibit any form of bias with regard to these demographic factors. This issue highlights the importance of transparently reporting the characteristics of study participants in order to assess the generalizability and potential biases of the findings. Therefore, using the proposed models for the diagnosis of COVID-19 infection in real-world applications, such as testing them on a private database of a local hospital, is a valuable next step in determining their clinical validity. This would involve collecting data from patients diagnosed with COVID-19, as well as from a control group of individuals, along with their demographic characteristics, and using this data to train and test the models. In this way, one can learn their clinical validity and improve them accordingly. Once the models have been validated, they can be used to aid in the diagnosis of COVID-19 in a clinical setting. This could involve integrating the models into existing electronic medical record systems, or developing a standalone diagnostic tool that can be used by healthcare providers.
4. Conclusions
In an effort to curb the spread of COVID-19, numerous deep learning-based models have recently been developed to detect patients with this disease as early as possible using chest X-ray images. However, most of these developed models are highly complex (heavyweight) and require a lot of computing power, making them unsuitable for devices with limited resources, such as smartphones. In recent times, devices like these have gained adoption everywhere, and thus they are being advocated to show great promise for the rapid diagnosis of COVID-19 infection. Motivated by the earlier discussion, two lightweight CNN-based models were proposed in this research to efficiently detect COVID-19 subjects from chest X-ray images. The proposed models are mainly 2-class (COVID-19 and Normal) and 3-class (COVID-19, Normal, and Viral Pneumonia) classification models, which were structured to contain learnable parameters not exceeding 600 K; hence, they are referred to as lightweight. Results from experiments on a sizable COVID-19 radiography dataset demonstrate that the proposed models achieved excellent performance in terms of accuracy, sensitivity, specificity, MCC, and AUC-ROC measures, without showing any indication of overfitting. For instance, we obtained an overall test accuracy rate of 98.55% in the binary classification and 96.83% in the multiclass classification tasks, which are really encouraging. Furthermore, our models require substantially lower computing power and memory resources than the existing CNN-based heavyweight models while maintaining comparable accuracy. Based on these results, we can conclude that our models may aid physicians in making meaningful diagnoses of COVID-19 infection and are suitable for implementation on devices with limited resources and power consumption.
In the future, we aim to apply some image preprocessing algorithms to the X-ray images to determine the extent of their impact on improving the proposed models’ performance. We also intend to use the proposed models in real-world applications, like testing them on the private database of a local hospital, so that we can learn their clinical validity and improve them accordingly.
CRediT authorship contribution statement
Haval I. Hussein: Conceptualization, Software, Investigation, Validation, Writing – original draft. Abdulhakeem O. Mohammed: Conceptualization, Methodology, Writing – review & editing. Masoud M. Hassan: Methodology, Software, Investigation, Validation, Writing – review & editing. Ramadhan J. Mstafa: Validation, Resources, Supervision, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Data availability
Data will be made available on request.
References
- Abdani S.R., Zulkifley M.A., Zulkifley N.H. A lightweight deep learning model for COVID-19 detection. IEEE Symposium on Industrial Electronics & Applications (ISIEA) 2020;2020:1–5. doi: 10.1109/ISIEA49364.2020.9188133. [DOI] [Google Scholar]
- Aggarwal P., Mishra N.K., Fatimah B., Singh P., Gupta A., Joshi S.D. COVID-19 image classification using deep learning. Advances, challenges and opportunities. 2022;144 doi: 10.1016/j.compbiomed.2022.105350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahrabi S.S., Scarpiniti M., Baccarelli E., Momenzadeh A. An accuracy vs. complexity comparison of deep learning architectures for the detection of covid-19 disease. Computation. 2021;9(1):1–20. doi: 10.3390/computation9010003. [DOI] [Google Scholar]
- Akter S., Shamrat F.M.J.M., Chakraborty S., Karim A., Azam S. Covid-19 detection using deep learning algorithm on chest X-ray images. Biology. 2021;10(11):1174. doi: 10.3390/biology10111174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alghamdi H.S., Amoudi G., Elhag S., Saeedi K., Nasser J. Deep learning approaches for detecting COVID-19 from Chest X-ray images: A survey. IEEE Access. 2021;9:20235–20254. doi: 10.1109/ACCESS.2021.3054484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali M.M., Paul B.K., Ahmed K., Bui F.M., Quinn J.M.W., Moni M.A. Heart disease prediction using supervised machine learning algorithms: Performance analysis and comparison. Computers in Biology and Medicine. 2021;136 doi: 10.1016/j.compbiomed.2021.104672. [DOI] [PubMed] [Google Scholar]
- Alshazly H., Linse C., Barth E., Martinetz T. Explainable COVID-19 detection using chest CT scans and deep learning. Sensors. 2021;21(2):1–22. doi: 10.3390/s21020455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolopoulos I.D., Aznaouridis S.I., Tzani M.A. Extracting possibly representative COVID-19 biomarkers from X-ray images with deep learning approach and image data related to pulmonary diseases. Journal of Medical and Biological Engineering. 2020;40:462–469. doi: 10.1007/s40846-020-00529-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolopoulos I.D., Mpesiana T.A. Covid-19: Automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Physical and Engineering Sciences in Medicine. 2020;43:635–640. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya A., Bhaik D., Kumar S., Thakur P., Sharma R., Pachori R.B. A deep learning based approach for automatic detection of COVID-19 cases using chest X-ray images. Biomedical Signal Processing and Control. 2022;71 doi: 10.1016/j.bspc.2021.103182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary P.K., Pachori R.B. FBSED based automatic diagnosis of COVID-19 using X-ray and CT images. Computers in Biology and Medicine. 2021;134 doi: 10.1016/j.compbiomed.2021.104454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung M., Bernheim A., Mei X., Zhang N., Huang M., Zeng X.…Shan H. CT imaging features of 2019 movel coronavirus. Radiology. 2020;295:202–207. doi: 10.1148/radiol.2020200230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Convolution operation. (2022). https://indoml.com/2018/03/07/student-notes-convolutional-neural-networks-cnn-introduction/.
- coronavirus worldometer. (2022). https://www.worldometers.info/coronavirus/.
- COVID-19 radiography database. (2022). https://www.kaggle.com/datasets/tawsifurrahman/covid19-radiography-database.
- Dhillon A., Verma G.K. Convolutional neural network: A review of models, methodologies and applications to object detection. Progress in Artificial Intelligence. 2020;9:85–112. doi: 10.1007/s13748-019-00203-0. [DOI] [Google Scholar]
- Fang L., Wang X. COVID-RDNet: A novel coronavirus pneumonia classification model using the mixed dataset by CT and X-rays images. Biocybernetics and Biomedical Engineering. 2022;42:977–994. doi: 10.1016/j.bbe.2022.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George G.S., Mishra P.R., Sinha P., Prusty M.R. COVID-19 detection on chest X-ray images using homomorphic transformation and VGG inspired deep convolutional neural network. Biocybernetics and Biomedical Engineering. 2023;43:1–16. doi: 10.1016/j.bbe.2022.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A., Katarya R. PAN-LDA: A latent Dirichlet allocation based novel feature extraction model for COVID-19 data using machine learning. Computers in Biology and Medicine. 2021;138 doi: 10.1016/j.compbiomed.2021.104920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A., Katarya R. Possibility of the COVID-19 third wave in India: Mapping from second wave to third wave. Indian Journal of Physics. 2023;97:389–399. doi: 10.1007/s12648-022-02425-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A., Katarya R. Anovel LDA-based framework to forecast COVID-19 trends. Proceedings of the International Conference on Innovative Computing & Communication (ICICC) 2021;2021:1–4. doi: 10.2139/ssrn.3833706. [DOI] [Google Scholar]
- Gupta A., Katarya R. COVID-19 cases prediction based on LSTM and SIR model using social media. Lecture Notes in Electrical Engineering. 2022;940:111–117. doi: 10.1007/978-981-19-4453-6_7. [DOI] [Google Scholar]
- Gupta, Anunay, Anjum, Gupta, S., & Katarya, R. (2021). InstaCovNet-19: A deep learning classification model for the detection of COVID-19 patients using chest X-ray. Applied Soft Computing Journal, 99, 106859. 10.1016/j.asoc.2020.106859. [DOI] [PMC free article] [PubMed]
- Haghanifar A., Majdabadi M.M., Choi Y., Deivalakshmi S., Ko S. COVID-CXNet: Detecting COVID-19 in frontal chest X-ray images using deep learning. Multimedia Tools and Applications. 2022;81:30615–30645. doi: 10.1007/s11042-022-12156-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan M.K., Alam M.A., Dahal L., Roy S., Wahid S.R., Elahi M.T.E.…Khanal B. Challenges of deep learning methods for COVID-19 detection using public datasets. Informatics in Medicine Unlocked. 2022;30 doi: 10.1016/j.imu.2022.100945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidari M., Mirniaharikandehei S., Khuzani A.Z., Danala G., Qiu Y., Zheng B. Improving the performance of CNN to predict the likelihood of COVID-19 using chest X-ray images with preprocessing algorithms. International Journal of Medical Informatics. 2020;144 doi: 10.1016/j.ijmedinf.2020.104284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertel R., Benlamri R. A deep learning segmentation-classification pipeline for X-ray-based COVID-19 diagnosis. Biomedical Engineering Advances. 2022;3 doi: 10.1016/j.bea.2022.100041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain E., Hasan M., Rahman M.A., Lee I., Tamanna T., Parvez M.Z. CoroDet: A deep learning based classification for COVID-19 detection using chest X-ray images. Chaos, Solitons & Fractals. 2021;110495 doi: 10.1016/j.chaos.2020.110495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ieracitano C., Mammone N., Versaci M., Varone G., Ali A., Armentano A.…Morabito F.C. A fuzzy-enhanced deep learning approach for early detection of Covid- 19 pneumonia from portable chest X-ray images. Neurocomputing. 2022;481:202–215. doi: 10.1016/j.neucom.2022.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalali S.M.J., Ahmadian M., Ahmadian S., Hedjam R., Khosravi A., Nahavandi S. X-ray image based COVID-19 detection using evolutionary deep learning approach. Expert Systems with Applications. 2022;201 doi: 10.1016/j.eswa.2022.116942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jyoti K., Sushma S., Yadav S., Kumar P., Pachori R.B., Mukherjee S. Automatic diagnosis of COVID-19 with MCA-inspired TQWT-based classification of chest X-ray images. Computers in Biology and Medicine. 2023;152 doi: 10.1016/j.compbiomed.2022.106331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katarya, R., Anjum, Gupta, A., Sachdeva, S., Dhamija, T., Gupta, S., Gupta, A., Kedia, P., & Rai, V. (2021). A review of various mathematical and deep learning based forecasting methods for COVID-19 pandemic. 7th International Conference on Advanced Computing and Communication Systems, ICACCS 2021, 874–878. 10.1109/ICACCS51430.2021.9441966.
- Katarya R., Nath G.A., Singhal D., Shukla A. Analysing the Twitter sentiments in COVID-19 using machine learning algorithms. IEEE International Conference on Advances in Computing, Communication and Applied Informatics, ACCAI. 2022;2022:1–8. doi: 10.1109/ACCAI53970.2022.9752511. [DOI] [Google Scholar]
- Khan A.I., Shah J.L., Bhat M.M. CoroNet: A deep neural network for detection and diagnosis of COVID-19 from chest x-ray images. Computer Methods and Programs in Biomedicine. 2020;196 doi: 10.1016/j.cmpb.2020.105581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M., Mehran M.T., Haq Z.U., Ullah Z., Naqvi S.R., Ihsan M., Abbass H. Applications of artificial intelligence in COVID-19 pandemic: A comprehensive review. Expert Systems with Applications. 2021;185 doi: 10.1016/j.eswa.2021.115695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Liu F., Yang W., Peng S., Zhou J. A survey of convolutional neural networks: Analysis, applications, and prospects. IEEE Transactions on Neural Networks and Learning Systems. 2021;1–21 doi: 10.1109/TNNLS.2021.3084827. [DOI] [PubMed] [Google Scholar]
- Luz E., Silva P., Silva R., Silva L., Guimarães J., Miozzo G.…Menotti D. Towards an effective and efficient deep learning model for COVID-19 patterns detection in X-ray images. Research on Biomedical Engineering. 2022;38:149–162. doi: 10.1007/s42600-021-00151-6. [DOI] [Google Scholar]
- Malik, D., Anjum, & Katarya, R. (2022). Comparative analysis by transfer learning of pre-trained models for detection of COVID-19 using chest X-ray images. Proceedings of the International Conference on Paradigms of Communication, Computing and Data Sciences, 549–557. 10.1007/978-981-16-5747-4_46.
- Minaee S., Kafieh R., Sonka M., Yazdani S., Soufi G.J. Deep-COVID: Predicting COVID-19 from chest X-ray images using deep transfer learning. Medical Image Analysis. 2020;65 doi: 10.1016/j.media.2020.101794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narin A., Kaya C., Pamuk Z. Automatic detection of coronavirus disease (COVID-19) using X-ray images and deep convolutional neural networks. Pattern Analysis and Applications. 2021;24:1207–1220. doi: 10.1007/s10044-021-00984-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak S.R., Nayak D.R., Sinha U., Arora V., Pachori R.B. Application of deep learning techniques for detection of COVID-19 cases using chest X-ray images: A comprehensive study. Biomedical Signal Processing and Control. 2021;64 doi: 10.1016/j.bspc.2020.102365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak S.R., Nayak D.R., Sinha U., Arora V., Pachori R.B. An efficient deep learning method for detection of COVID-19 infection using chest X-ray images. Diagnostics. 2023;13(1):1–17. doi: 10.3390/diagnostics13010131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin M.E. Deep learning-based approach for detecting COVID-19 in chest X-rays. Biomedical Signal Processing and Control. 2022;78 doi: 10.1016/j.bspc.2022.103977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewariya M., Katarya R. Parameter based literature survey of COVID-19 mortality dynamics using machine learning techniques. International Conference on Recent Trends on Electronics, Information, Communication and Technology, RTEICT. 2021;2021:141–145. doi: 10.1109/RTEICT52294.2021.9573980. [DOI] [Google Scholar]
- Thakur, S., Kasliwal, Y., Kothambawala, T., & Katarya, R. (2022). A Study on pulmonary image screening for the detection of COVID-19 using convolutional neural networks. In Data Engineering for Smart Systems. Lecture Notes in Networks and Systems (Vol. 238). 10.1007/978-981-16-2641-8_44.
- Ukwandu O., Hindy H., Ukwandu E. An evaluation of lightweight deep learning techniques in medical imaging for high precision COVID-19 diagnostics. Healthcare Analytics. 2022;2 doi: 10.1016/j.health.2022.100096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Lin Z.Q., Wong A. COVID-Net: A tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images. Scientific Reports. 2020;10:19549. doi: 10.1038/s41598-020-76550-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., Wenjie T. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA - Journal of the American Medical Association. 2020;323:1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita R., Nishio M., Do R.K.G., Togashi K. Convolutional neural networks: An overview and application in radiology. Insights into Imaging. 2018;9:611–629. doi: 10.1007/s13244-018-0639-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zebin T., Rezvy S. COVID-19 detection and disease progression visualization: Deep learning on chest X-rays for classification and coarse localization. Applied Intelligence. 2021;51:1010–1021. doi: 10.1007/s10489-020-01867-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.