Abstract
Cryo-Electron Microscopy (cryo-EM) has emerged as a key technology to determine the structure of proteins, particularly large protein complexes and assemblies in recent years. A key challenge in cryo-EM data analysis is to automatically reconstruct accurate protein structures from cryo-EM density maps. In this review, we briefly overview various deep learning methods for building protein structures from cryo-EM density maps, analyze their impact, and discuss the challenges of preparing high-quality data sets for training deep learning models. Looking into the future, more advanced deep learning models of effectively integrating cryo-EM data with other sources of complementary data such as protein sequences and AlphaFold-predicted structures need to be developed to further advance the field.
Keywords: cryo-electron microscopy (cryo-EM), protein structure, machine learning, deep learning
Graphical Abstract
1. Introduction
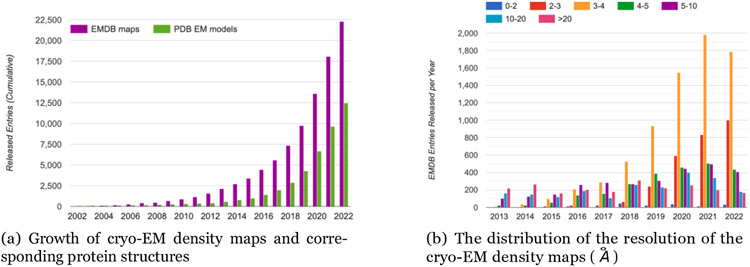
Cryo-EM is revolutionizing structural biology due to its unique capability of determining the structures of large protein complexes and assemblies. The atomic-resolution structure determination for proteins enabled by cryogenic electron microscopy (cryo-EM) [3], allows us to understand the complex biological processes carried out by proteins as well as to identify potential therapeutic protein targets for drug discovery. However, reconstructing de novo protein structures from high-resolution (~ 3 - 4 Å) cryo-EM density maps, which accounts for a large portion of cryo-EM density maps deposited currently in the EMDB [2], is time-consuming and challenging when homologous template structures for target proteins are not available. For instance, as shown in Figure 1, in the current year 2022, only about 12,500 out of 22,300 density maps of high-resolutions deposited to EMDB have a complete atomic structure available in Protein Data Bank (PDB) [40].
Figure 1:
The growth of cryo-EM density maps and cryo-EM-derived protein structures and the distribution of the resolution of the density maps. The statistics was obtained from EMDataResource [2], an unified data resource for 3-Dimension electron microscopy (3DEM) on 2022-09-14.
Accurately reconstructing protein structures from cryo-EM maps is a challenging process because the data is often noisy and incomplete and target protein structures can be large and complex. Traditional methods based on energy optimization such as EM-Fold [23], Gorgon [24], Rosetta [25], Pathwalking [26], MAINMAST [27, 28], VESPER [51], and Phenix [29] have made valuable progress in reconstructing protein structures from cryo-EM density maps. These methods rely on extensive physics-based or statistical potential-based optimization algorithms that require high computational resources. These methods often need manual intervention and trials to extract features from the cryo-EM density maps to obtain accurate reconstruction of protein structure.
A different strategy to automatically determine protein structures from cryo-EM density maps is to use the data-driven machine learning approach [44], a kind of artificial intelligence (AI) technology, to directly learn a mapping from cryo-EM density maps to protein structures from the large amount of known cryo-EM data and their corresponding protein structures (i.e., labels). Early AI methods in the field are based on shallow machine learning techniques such as k-nearest neighbor, support-vector machines, or k-means clustering techniques. These methods such as RENNSH [30], SSELearner [31], and Pathwalking [26] are able to identify only secondary structures or simplified backbone structures and often are unable to achieve the optimal solution.
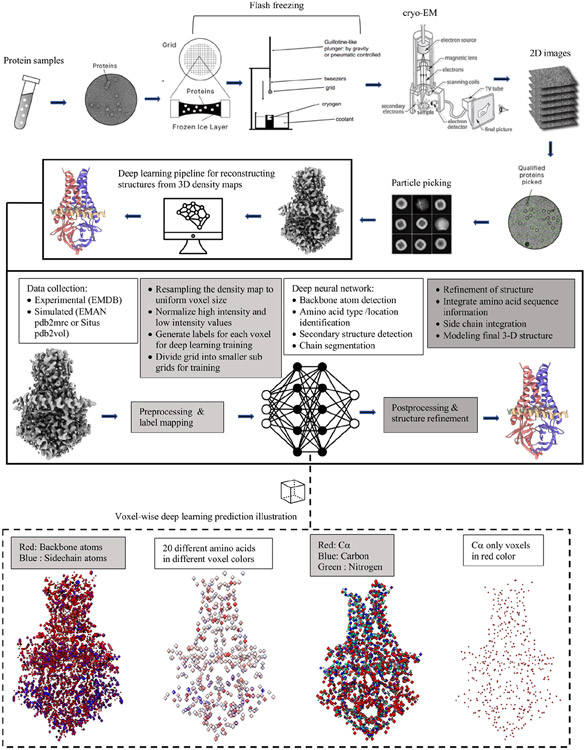
To overcome the challenges of the traditional optimization methods and early machine learning methods, deep learning methods [45] have been developed to automatically reconstruct three-dimensional (3D) protein structures from cryo-EM density maps with significant success in recent years (see Figure 2 for a summary of a general cryo-EM protein structure determination pipeline powered by deep learning). In this article, we review the recent development of deep learning technology in the field, analyze their impacts, investigate the challenging issues in preparing data to train deep learning models, and discuss some new trends to further advance the field.
Figure 2:
A summary of a cryo-EM density map generation and protein structure reconstruction pipeline powered by deep learning. The density map (EMD-22898) illustrated in the figure is for SARS-CoV-2 ORF3a [39]. PDB ID: 7KJR.
2. Deep learning reconstruction of protein structures from cryo-EM density maps
Deep learning, also called deep neural network, is currently the most powerful machine learning method of predicting the properties of an object from the input data describing the object. It has achieved great success in many fields including a recent major breakthrough in predicting protein structure from sequence by AlphaFold [1]. Compared to other machine learning methods, deep learning has a unique capability of extracting informative features for pattern recognition from raw data automatically, making it suitable for reconstructing protein structures from raw density maps in which only a large amount of numbers rather than informative features are available.
It is worth noting that deep learning has been applied to almost all the areas of cryo-EM data analysis [35, 32, 19, 20, 21, 22, 38] from sample preparation, particle picking, density map denoising, and to the final step of 3-D structure determination. Due to the space limit, this review is focused on the last step of cryo-EM data analysis - reconstructing protein structures from density maps. The deep learning architectures designed for this task and how to prepare data to train them are discussed in the two subsections below.
2.1. Deep learning architectures for reconstructing protein structures from cryo-EM density maps
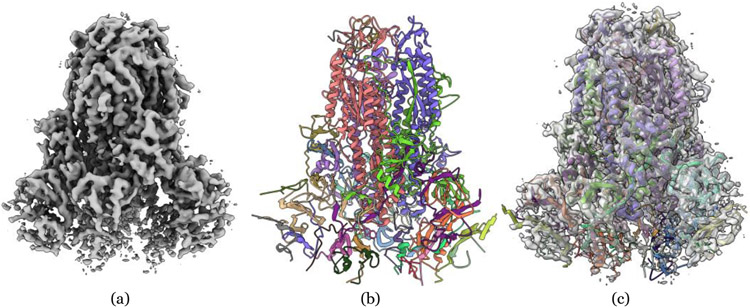
Deep learning methods for inferring protein structures from cryo-EM density maps can be classified into different categories based on the neural network architectures, for example, convolutional neural network (CNN) [33], U-Net [34, 43], graph convolutional network (GCN) [41], and long- and short-term memory network (LSTM) [42] they use and the output (e.g., 3D structure and secondary structure) they generate from density map input. Early deep learning methods aimed to identity secondary structures from low- and medium-resolution density maps [11]. As more and more high-resolution density maps became available [3], recent deep learning methods targeted at directly reconstruct 3D backbone structures (i.e., locations of carbon and nitrogen atoms on the protein backbone) and even full-atom 3D structures (i.e., locations of all/most heavy atoms and amino acid identity/type) from density maps [10, 7, 14, 15, 16]. An example of deep learning reconstruction of protein structure from cryo-EM density map is showed in Figure 3.
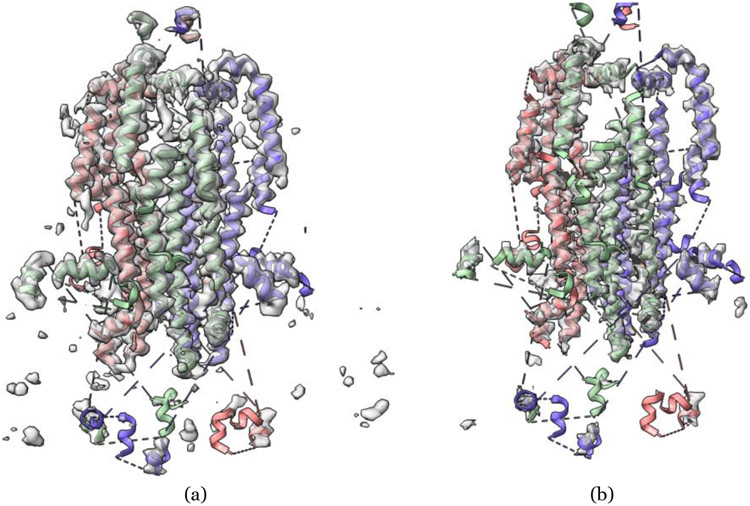
Figure 3:
An example of reconstructing a structure from the cryo-EM density map of SARS-CoV spike gycoprotein by deep learning. (a) Density map of SARS-CoV spike glycoprotein [48] (EMD-6732) in resolution of 3.8 Å at recommended contour level of 0.06 (11.0 σ). (b) The structure reconstructed from EMD-6732 by a deep learning method - DeepTracer. The RMSD is 1.023 Å with respect to the ground truth structure (PDB ID: 5XLR). (c) The overlay of the density map and reconstructed structure at 0.5 transparency level by UCSF ChimeraX [49].
One of the most widely used deep learning architectures of obtaining protein structural information from density maps is convolution neural network (CNN). CNNs use a mathematical operation known as convolution to extract features from spatially organized data such as a 2D-image or 3D density map to predict the properties of the data (e.g., classifying voxels in a density map into amino acid types). Several CNN methods (mostly 3D-CNN architecture) including Generator [7], Emap2sec [8], AAnchor [9], CNN Based [11], Cascaded-CNN [10], and CR-I-TASSER (mostly 3D CNN) [15] have been developed to determine secondary structures [8, 11], backbone-/full-atom 3D structures [15, 7, 9] or both from cryo-EM density maps [10]. Cascaded-CNN is the first deep learning de novo method of directly reconstructing 3D structures of proteins from cryo-EM density maps, even though it focuses on building backbone structures. CR-I-TASSER combines the 3D-CNN prediction from cryo-EM maps and an advanced protein structure prediction method - I-TASSER [46] to build full-atom protein structures.
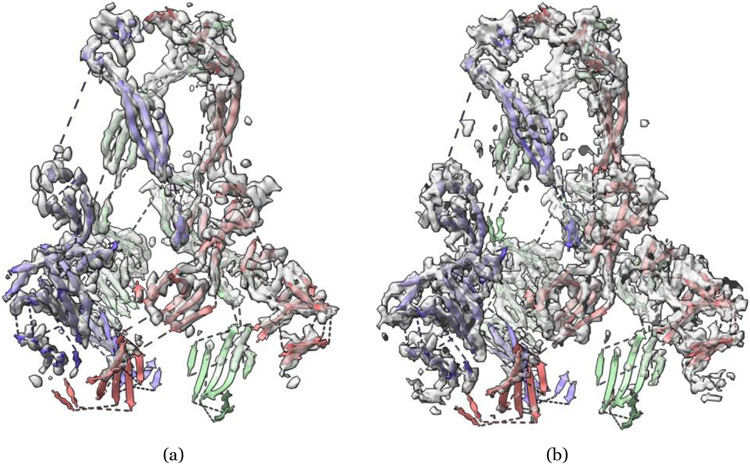
Another widely used convolutional neural network architecture in the field is U-Net [34], originally designed for biomedical image classification and segmentation tasks. U-Net consists of a series of convolution-based down-sampling layers to condense the input images into smaller dimensions and a series of convolution-based up-sampling counterpart layers to reconstruct the data of the same dimension as in the down-sampling process to classify/segment pixels in the input images. Compared to the standard CNN architectures, U-Nets can be more effective in extracting multi-level abstract representations of the data through the down-sampling and up-sampling processes. The 2D U-Net architecture has been generalized to 3D U-Net architectures in Haruspex [12] and EMNUSS [17] to detect secondary structures from cryo-EM density maps (e.g., Figure 4, and 5), and in DeepTracer [13] and EMBuild [16] to reconstruct 3D protein structures from cryo-EM density maps. DeepTracer has been successfully applied to reconstruct the structures of some SARS-CoV proteins from cryo-EM density maps (e.g., Figure 3).
Figure 4:
An example of secondary structure annotation in cryo-EM density map of SARS-CoV spike gycoprotein [48] (EMD-6732) by deep learning. PDB ID: 5XLR. (a) Haruspex [12] predicted strands in transparent gray overlapped with deposited PDB structure strands. (b) EMNUSS [17] predicted strands in transparent gray overlapped with deposited PDB structure strands.
Figure 5:
An example of secondary structure annotation in cryo-EM density map of SARS-CoV spike gycoprotein [48] (EMD-6732) by deep learning. PDB ID: 5XLR. (a) Haruspex [12] predicted helices in transparent gray overlapped with deposited PDB structure helices. (b) EMNUSS [17] predicted helices in transparent gray overlapped with deposited PDB structure helices.
In addition to CNN and U-Net, other deep learning architectures such as graph convolutional networks (GCN) and long- and short-term memory network (LSTM) have also been used with CNN to reconstruct protein structures from cryo-EM density maps [7]. A summary of different deep learning-based methods, their function (e.g., input and output) and availability is presented in Table 1.
Table 1:
Summary of deep learning based methods for protein structure reconstruction from cryo-EM density maps.
| Methods | Architecture | Function | Open source |
|---|---|---|---|
| Structure Generator[7] | 3-D CNN, GCN, Bidirectional LSTM | First use 3-D CNN to identify amino acids and their rotameric identities in an EM map and then GCN and LSTM to build protein structures | ✓ |
| Emap2sec[8] | 3-D CNN | Take voxel cubes as input to identify secondary structures of protein | ✓ |
| AAnchor[9] | 3-D CNN | Take in voxel cubes to identify amino acid types and locations | ✓ |
| A CNN Based Method[11] | 3-D CNN | Take in voxel cubes to detect secondary structures of protein from background | × |
| CascadedCNN[10] | Cascaded 3-D CNN | Take in voxel cubes to identify Cα atoms of protein backbone and secondary structures to generate 3D protein structures | ✓ |
| Haruspex[12] | 3-D U-Net | Take in voxel cubes to predict the probabilities of 4 different classes; α-helix, β-sheet, nucleotide, or unassigned to assign secondary structures | ✓ |
| DeepTracer[13] | 3-D U-Net | Take in voxel cubes to identify the location of backbone atoms, secondary structures and amino acid types simultaneously to build 3D structure | × |
| DeepTracer ID[14] | DeepTracer (3-D U-Net) and pre-calculated AlphaFold2 protein library | Use DeepTracer to generate an initial 3D protein structure to search AlphaFold2DB to identify similar structural hits for refinement | × |
| CR-I-TASSER [15] | 3-D CNN, I-TASSER | Predict Cα using 3-D CNN for selecting structural templates for I-TASSER to generate 3D protein structure | ✓ |
| EMBuild [16] | 3-D U-Net++, AlphaFold | Integrate AlphaFold structure pre-diction, FFT-based global fitting, domain-based semi-flexible refinement, and graph-based iterative assembling with main-chain probability maps predicted by U-Net++ to build 3D protein structure | ✓ |
| EMNUSS [17] | 3-D U-Net++ | Take in voxel cubes to identify secondary structures of protein | ✓ |
| ModelAngelo [18] | Graph Neural Network | Refines geometry of protein chains and classifies amino acid for each nodes | × |
Inspired by the recent breakthrough in developing deep learning methods of predicting protein structures from sequences such as AlphaFold [1] and RoseTTAFold [5], a new trend is to integrate deep learning methods of reconstruct protein structures from cryo-EM density maps with the advanced computational (e.g., deep learning) methods of predicting protein structures from sequences to obtain more accurate structural models. For instance, DeepTracer ID [14] first uses DeepTracer to build an initial structure from cryo-EM density maps and then search the structure against a database of AlphaFold-predicted structures to identify similar structural hits to enhance the reconstructed structure. EMBuild [16] combines the structures reconstructed from cryo-EM maps, AlphaFold-predicted structural models and other protein structural refinement methods to construct accurate structures for protein complexes. ModelAngelo [18] refines the geometry of protein chains by combining information extracted from cryo-EM data, prior knowledge of protein geometries, and amino acid sequence data. DeepProLigand [4] integrates the protein structural models reconstructed from cryo-EM density maps by DeepTracer with the known template structures containing ligands to model protein-ligand interaction, which was ranked first in the ligand prediction in 2021 EMDataResource Ligand Model Challenge.
3. Data preparation for training deep learning methods to reconstructing protein structures from cryo-EM density maps
3.1. Cryo-EM density map data collection
Collecting a sufficient amount of high-quality data to train and test deep learning models is critical for any deep learning task. The common way to acquire the experimental cryo-EM density maps is through the Electron Microscopy Data Bank [2]. An alternative approach employed by some methods such as Cascaded-CNN [10] and SSELearner [31] is to simulate the density map from the PDB protein structure. Cascaded-CNN applies pdb2mrc from EMAN2 package [50], and VESPER uses pdb2vol from Situs package [52] to generate the simulated maps. However, simulated maps lack complex noise, missing density values, and experimental artifacts which can arise from particle alignment errors, interaction of electron beam with the atoms, or movement of atoms during image capture. Therefore, the deep learning models trained on simulated maps may not work as expected on very noisy experimental data. To address the problem, CR-I-TASSER, EMNUSS and Emap2sec employs a hybrid training approach that uses both simulated maps and experimental maps in the training and validation process.
3.2. Training data preprocessing
Prior to using the cryo-EM density map to train deep learning models, it is generally necessary to normalize and standardize the data to make them suitable for deep learning as shown by Cascaded-CNN and DeepTracer, which perform data grid resampling, density value normalization, and grid division. These preprocessing steps ensure the uniformity among density maps and help deep learning models to extract features and recognize patterns more easily. During the grid division, the 3D cryo-EM is splitted into the cubes of a specific size (e.g., 64 × 64 × 64 Å3 by Cascaded-CNN and DeepTracer, 50 × 50 × 50 Å3 by CR-I-TASSER, 40 × 40 × 40 Å3 by Haruspex, and 11 × 11 × 11 Å3 by Emap2sec and AAnchor). Each of these cubes is then processed by the deep learning method to classify the voxels into the targeted classes such as amino acid types (identities) and secondary structures.
4. Future directions
Deep learning has made a significant impact on protein structure reconstruction from cryo-EM density maps. However, the field is still in the early stage of development. The latest deep learning technology such as graph neural networks [53] and attention mechanisms [47] have not been extensively used in the field. While CNNs and U-Nets based on convolution are currently the most used methods for structure reconstruction, they have some short-coming for 3D structural modeling. CNNs are translation-equivariant, but not fully rotation invariant that is desirable for 3D structure analysis. Moreover, the convolution mechanism propagates message in the constrained local receptive field, which is not as effective as the attention mechanism [47] that can leverage all the input information by automatically weighting the input features according to their relevance as demonstrated by the remarkable success of AlphaFold2 in protein structure prediction. More sophisticated deep learning models like attention-based Transformer models [36], 3D-equivariant graph neural networks [37], and AlphaFold2-like deep learning models need to be developed to better use cryo-EM data to improve reconstruction accuracy.
Another important direction is to use deep learning to integrate cryo-EM data with multiple other sources of complementary data such as protein structural models predicted from sequences, structural templates in the Protein Data Bank, and protein sequences to more accurately reconstruct protein structures from noisy density maps that often miss the density values of some atoms. The current integration process is limited to shallow data combination. For instance, DeepTracer ID uses AlphaFold models to refine the structural models predicted from structural models reconstructed from deep learning. More comprehensive, end-to-end deep learning models to combine multiple sources of data to generate accurate final protein structures can be developed to automatically and accurately reconstruct protein structures from the data.
Moreover, it is important to integrate cryo-EM based deep learning methods of reconstructing protein structures with the advanced methods developed in the field of protein structure prediction. The structural models directly reconstructed from cryo-EM data by deep learning generally have correct overall topology, but the reconstructed models may not satisfy physicochemical restraints such as bond length and bond angles and not have all the molecular details (e.g., the precise location of all side chain atoms) [10, 4]. Linking the atoms of amino acids identified from the density maps into full peptide chains consistent with protein sequences and physical-chemical restraints is still challenging. However, the modeling techniques such as protein structure refinement and molecular dynamics to fix these problems have been established for protein structure prediction [1]. Some methods such as CR-I-TASSER have started to integrate the two kinds of technologies. More synergistic integration of the two are needed to generate high-quality realistic protein structures from cryo-EM data.
The development of high-quality deep learning models to reconstruct protein structures from cryo-EM density maps critically depends on the availability of sufficient high-quality training data. Although experimental cryo-EM data and its related ground truth structure are freely accessible through EMDB [2] and RCSB PDB [40], these datasets still need to be pre-processed and labeled before they can be used for deep learning training. Curating a large amount of high-quality training and test data is challenging and time consuming, but often receives little attention. Currently, there are few well-curated experimental cryo-EM data sets publicly available for training and evaluating deep learning models in the field. Therefore, more effort needs to be devoted to creating such data sets and make them to publicly available for the community to use.
5. Conclusion
A number of useful deep learning models have been developed to reconstruct protein structures from cryo-EM density maps, demonstrating deep learning is a promising technology to further push the frontier of applying cryo-EM technology to determine protein structures. As the deep learning field is evolving very fast, many more state-of-the-art deep learning architectures (e.g., AlphaFold2-like models and transformers) have yet to be applied to further advance the emerging field. More sophisticated deep learning methods need to be developed to seemlessly integrate cryo-EM data with other complementary data such as predicted protein structures, protein sequences, and template structures to further improve cryo-EM-based structure determination. A synergistic integration of cryo-EM based protein structure determination techniques and latest protein structure prediction techniques is also important for generating highly accurate native-like protein structures. To speed up the development, more effort is need to create a large amount of high-quality cryo-EM training and test data for the community to use.
Highlights.
Deep learning is a promising technique for efficient, automatic, and accurate reconstruction of protein structures from cryo-EM density maps
Advanced convolutional neural networks and U-Nets have been successfully applied to reconstruct protein structures from high-resolution cryo-EM density maps
Creating high-quality cryo-EM data sets for training and testing deep learning methods is important and there is a significant need of curating such data sets to facilitate the development of deep learning methods
Better structure reconstruction can be obtained by combining AlphaFold predicted structure models and cryo-EM data and by integrating cryo-EM based structure determination techniques and protein structure prediction techniques.
More advanced deep learning architectures and better integration of multiple sources of complementary data are needed to advance the field
Acknowledgements
This work was supported in part by Department of Energy grants (DE-AR0001213, DE-SC0020400, and DE-SC0021303), two NSF grants (DBI1759934 and IIS1763246), and NIH grants (R01GM093123 and R01GM146340).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
The authors declare that there is no conflict of interest.
References
Papers of particular interest, published within the period of review, have been highlighted as:
* of special interest
** of outstanding interest
- [1] **. Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Z′ıdek A, Potapenko A, et al. : Highly accurate protein structure prediction with AlphaFold. Nature 2021, 10.1038/S41586-021-03819-2. Highly accurate deep neural network based system that predicts protein’s 3D structure form its amino acid sequence.
- [2].Lawson Catherine L., Patwardhan Ardan, Baker Matthew L., Hryc Corey, Garcia Eduardo Sanz, Hudson Brian P., Lagerstedt Ingvar, Ludtke Steven J., Pintilie Grigore, Sala Raul, Westbrook John D., Berman Helen M., Kleywegt Gerard J., Chiu Wah, EMDataBank unified data resource for 3DEM, Nucleic Acids Research, Volume 44, Issue D1, 4 January 2016, Pages D396–D403, 10.1093/nar/gkv1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Werner Kühlbrandt: The Resolution Revolution. Science 2014, 10.1126/science.1251652 , https://www.science.org/doi/abs/10.1126/science.1251652S41586-021-03819-2 [DOI] [Google Scholar]
- [4].Giri N, Cheng J (2022). A Deep Learning Bioinformatics Approach to Modeling Protein-Ligand Interaction with cryo-EM Data in 2021 Ligand Model Challenge. bioRxiv, doi: 10.1101/2022.05.27.493799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Baek M, DiMaio F, Anishchenko I, Dauparas J, Ovchinnikov S, Lee GR, … Baker D (2021). Accurate prediction of protein structures and interactions using a three-track neural network. Science, 373(6557), 871–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].He J, Huang SY (2021). EMNUSS: a deep learning framework for secondary structure annotation in cryo-EM maps. Briefings in bioinformatics, 22(6), bbab156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Li Po-Nan, de Oliveira Saulo HP, Soichi Wakatsuki, and van den Bedem Henry. ”Sequence-guided protein structure determination using graph convolutional and recurrent networks.” In 2020 IEEE 20th international conference on bioinformatics and bioengineering (BIBE), pp. 122–127. IEEE, 2020. [Google Scholar]
- [8].Subramaniya Maddhuri Venkata, Raghavendra Sai, Terashi Genki, and Kihara Daisuke. ”Protein secondary structure detection in intermediate-resolution cryo-EM maps using deep learning.” Nature methods 16, no. 9 (2019): 911–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Rozanov Mark, and Wolfson Haim J.. ”AAnchor: CNN guided detection of anchor amino acids in high resolution cryo-EM density maps.” In 2018 IEEE international conference on bioinformatics and biomedicine (BIBM), pp. 88–91. IEEE, 2018. [Google Scholar]
- [10] **. Si Dong, Moritz Spencer A., Pfab Jonas, Hou Jie, Cao Renzhi, Wang Liguo, Wu Tianqi, and Cheng Jianlin. ”Deep learning to predict protein backbone structure from high-resolution cryo-EM density maps.” Scientific reports 10, no. 1 (2020): 1–22 Among the first deep learning methods that accurately predict Cα positions along the protein’s backbone from cryo-EM density maps automatically.
- [11].Li Rongjian, Si Dong, Zeng Tao, Ji Shuiwang, and He Jing. ”Deep convolutional neural networks for detecting secondary structures in protein density maps from cryo-electron microscopy.” In 2016 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), pp. 41–46. IEEE, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12] *. Mostosi Philipp, Schindelin Hermann, Kollmannsberger Philip, and Thorn Andrea. ”Hamspex: a neural network for the automatic identification of oligonucleotides and protein secondary structure in cryo-electron microscopy maps.” Angewandte Chemie International Edition 59, no. 35 (2020): 14788–14795. Identifies secondary structures and nucleotides with high precision and recall.
- [13] **. Pfab Jonas, Nhut Minh Phan, and Dong Si. ”DeepTracer for fast de novo cryo-EM protein structure modeling and special studies on CoV-related complexes.” Proceedings of the National Academy of Sciences 118, no. 2 (2021): e2017525118. Builds 3D protein structure automatically and accurately from cryo-EM and amino acid sequence.
- [14] *. Chang Luca, Wang Fengbin, Connolly Kiernan, Meng Hanze, Su Zhangli, Virginija Cvirkaite-Krupovic Mart Krupovic, Egelman Edward H., and Si Dong. ”DeepTracer ID: De Novo Protein Identification from Cryo-EM Maps.” bioRxiv (2022). Predicts backbone structures using DeepTracer and searches them against AlphaFoldDB to refine the models.
- [15].Zhang Xi, Zhang Biao, Freddolino Peter L., and Zhang Yang. ”CR-I-TASSER: assemble protein structures from cryo-EM density maps using deep convolutional neural networks.” Nature Methods 19, no. 2 (2022): 195–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].He Jiahua, Lin Peicong, Chen Ji, Cao Hong, and Huang Sheng-You. ”Model building of protein complexes from intermediate-resolution cryo-EM maps with deep learning-guided automatic assembly.” Nature Communications 13, no. 1 (2022): 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].He Jiahua, and Huang Sheng-You. ”EMNUSS: a deep learning framework for secondary structure annotation in cryo-EM maps.” Briefings in bioinformatics 22, no. 6 (2021): bbab156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18] *.Jamali Kiarash, Kimanius Dari, and Scheres Sjors. ”ModelAngelo: Automated Model Building in Cryo-EM Maps.” arXiv preprint arXiv:2210.00006 (2022). [Google Scholar]
- [19] *. Zhong Ellen D., Bepler Tristan, Berger Bonnie, and Davis Joseph H.. ”CryoDRGN: reconstruction of heterogeneous cryo-EM structures using neural networks.” Nature methods 18, no. 2 (2021): 176–185. Classifies particle images in cryo-EM using variational autoencoder-decoder architecture.
- [20].Chen Muyuan, and Ludtke Steven J.. ”Deep learning-based mixed-dimensional Gaussian mixture model for characterizing variability in cryo-EM.” Nature methods 18, no. 8 (2021): 930–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lei Houchao, and Yang Yang. ”CDAE: a cascade of denoising autoencoders for noise reduction in the clustering of single-particle cryo-EM images.” Frontiers in genetics 11 (2021): 627746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kimanius Dari, Zickert Gustav, Nakane Takanori, Adler Jonas, Lunz Sebastian, Schönlieb C-B, Ö ktem Ozan, and Scheres Sjors HW. ”Exploiting prior knowledge about biological macromolecules in cryo-EM structure determination.” IUCrJ 8, no. 1 (2021): 60–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lindert Steffen, Nathan Alexander, Nils Wötzel, Mert Karaka**s, Phoebe L. Stewart, and Jens Meiler. ”EM-fold: de novo atomic-detail protein structure determination from medium-resolution density maps.” Structure 20, no. 3 (2012): 464–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Baker Matthew L., Abeysinghe Sasakthi S., Schuh Stephen, Coleman Ross A., Abrams Austin, Marsh Michael P., Hryc Corey F., Ruths Troy, Chiu Wah, and Ju Tao. ”Modeling protein structure at near atomic resolutions with Gorgon.” Journal of structural biology 174, no. 2 (2011): 360–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].DiMaio Frank, Andrew Leaver-Fay Phil Bradley, Baker David, and André Ingemar. ”Modeling symmetric macromolecular structures in Rosetta3.” PloS one 6, no. 6 (2011): 620450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Chen Muyuan, Baldwin Philip R., Ludtke Steven J., and Baker Matthew L.. ”De Novo modeling in cryo-EM density maps with Pathwalking.” Journal of structural biology 196, no. 3 (2016): 289–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Terashi Genki, Kagaya Yuki, and Kihara Daisuke. ”MAINMASTseg: automated map segmentation method for cryo-EM density maps with symmetry.” Journal of chemical information and modeling 60, no. 5 (2020): 2634–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Terashi Genki, and Kihara Daisuke. ”De novo main-chain modeling for EM maps using MAINMAST.” Nature communications 9, no. 1 (2018): 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Liebschner Dorothee, Afonine Pavel V., Baker Matthew L., Bunkãczi Gábor, Chen Vincent B., Croll Tristan I., Hintze Bradley et al. ”Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix.” Acta Crystallo-graphica Section D: Structural Biology 75, no. 10 (2019): 861–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Ma Lingyu, Reisert Marco, and Burkhardt Hans. ”RENNSH: A Novelα-Helix Identification Approach for Intermediate Resolution Electron Density Maps.” IEEE/ACM Transactions on Computational Biology and Bioinformatics 9, no. 1 (2011): 228–239. [DOI] [PubMed] [Google Scholar]
- [31].Si Dong, Ji Shuiwang, Nasr Kamal Al, and Jing He. ”A machine learning approach for the identification of protein secondary structure elements from electron cryo-microscopy density maps.” Biopolymers 97, no. 9 (2012): 698–708. [DOI] [PubMed] [Google Scholar]
- [32].Gupta Harshit, Michael T. McCann, Laurene Donati, and Michael Unser. ”CryoGAN: a new reconstruction paradigm for single-particle cryo-EM via deep adversarial learning.” IEEE Transactions on Computational Imaging 7 (2021): 759–774. [Google Scholar]
- [33].Rawat Waseem, and Wang Zenghui. ”Deep convolutional neural networks for image classification: A comprehensive review.” Neural computation 29, no. 9 (2017): 2352–2449. [DOI] [PubMed] [Google Scholar]
- [34] *. Ronneberger Olaf, Fischer Philipp, and Brox Thomas. ”U-net: Convolutional networks for biomedical image segmentation.” In International Conference on Medical image computing and computer-assisted intervention, pp. 234–241. Springer, Cham, 2015. U-Net, a widely used architecture to classify segment pixels in medical images.
- [35] *. Si Dong, Nakamura Andrew, Tang Runbang, Guan Haowen, Hou Jie, Firozi Ammaar, Cao Renzhi, Hippe Kyle, and Zhao Minglei. ”Artificial intelligence advances for de novo molecular structure modeling in cryo-electron microscopy.” Wiley Interdisciplinary Reviews: Computational Molecular Science 12, no. 2 (2022): e1542 A systematic review for AI methods in cryo-EM, covering implementation of AI in different stages of cryo-EM workflow.
- [36] **. Vaswani Ashish, Shazeer Noam, Parmar Niki, Uszkoreit Jakob, Jones Llion, Gomez Aidan N., Kaiser Łukasz, and Polosukhin Illia. ”Attention is all you need.” Advances in neural information processing systems 30 (2017). Deep learning model that uses self-attention module to learn and identify relationships in data. Originally used in the fields of natural language processing and computer vision.
- [37] **. Fuchs Fabian, Worrall Daniel, Fischer Volker, and Welling Max. ”Se (3)-transformers: 3d roto-translation equivariant attention networks.” Advances in Neural Information Processing Systems 33 (2020): 1970–1981. SE(3)-Transformer, a variant of the self-attention module for 3D point clouds, which is equivariant under continuous 3D roto-translations.
- [38].Al-Azzawi Adil, Ouadou Anes, Max Highsmith, Duan Ye, Tanner John J., and Cheng Jianlin. ”DeepCryoPicker: fully automated deep neural network for single protein particle picking in cryo-EM.” BMC bioinformatics 21, no. 1 (2020): 1–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kern David M et al. “Cryo-EM structure of SARS-CoV-2 ORF3a in lipid nanodiscs.” Nature structural and molecular biology vol. 28,7(2021): 573–582. doi: 10.1038/s41594-021-00619-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Stephen K Burley Charmi Bhikadiya, Bi Chunxiao et al. RCSB Protein Data Bank: powerful new tools for exploring 3D structures of biological macromolecules for basic and applied research and education in fundamental biology, biomedicine, biotechnology, bioengineering and energy sciences, Nucleic Acids Research, Volume 49, Issue D1, 8 January 2021, Pages D437–D451, 10.1093/nar/gkaa1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kipf Thomas N., and Welling Max. ”Semi-supervised classification with graph convolutional networks.” arXiv preprint arXiv:160902907. (2016). [Google Scholar]
- [42].Sherstinsky Alex. ”Fundamentals of recurrent neural network (RNN) and long short-term memory (LSTM) network.” Physica D: Nonlinear Phenomena 404 (2020): 132306. [Google Scholar]
- [43] *. Zhou Zongwei, Md Mahfuzur Rahman Siddiquee, Nima Tajbakhsh, and Jianming Liang. ”Unet++: A nested u-net architecture for medical image segmentation.” In Deep learning in medical image analysis and multimodal learning for clinical decision support, pp. 3–11. Springer, Cham, 2018. Nested U-Net, also known as U-Net ++, architecture that performs better than vanilla U-Net in segmentation tasks.
- [44].Greener Joe G., Kandathil Shaun M., Moffat Lewis, and Jones David T.. ”A guide to machine learning for biologists.” Nature Reviews Molecular Cell Biology 23, no. 1 (2022): 40–55. [DOI] [PubMed] [Google Scholar]
- [45].Esteva Andre, Chou Katherine, Yeung Serena, Naik Nikhil, Madani Ali, Mottaghi Ali, Liu Yun, Topol Eric, Dean Jeff, and Socher Richard. ”Deep learning-enabled medical computer vision.” NPJ digital medicine 4, no. 1 (2021): 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Yang Jianyi, Yan Renxiang, Roy Ambrish, Xu Dong, Poisson Jonathan, and Zhang Yang. ”The I-TASSER Suite: protein structure and function prediction.” Nature methods 12, no. 1 (2015): 7–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Guo Meng-Hao, Xu Tian-Xing, Liu Jiang-Jiang, Liu Zheng-Ning, Jiang Peng-Tao, Mu Tai-Jiang, Zhang Song-Hai, Martin Ralph R., Cheng Ming-Ming, and Hu Shi-Min. ”Attention mechanisms in computer vision: A survey.” Computational Visual Media (2022): 1–38. [Google Scholar]
- [48].Gui M, Song W, Zhou H, Xu J, Chen S, Xiang Y, Wang X. Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell Res. 2017. Jan;27(1):119–129. doi: 10.1038/cr.2016.152. Epub 2016 Dec 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Pettersen Eric F., Goddard Thomas D., Huang Conrad C., Meng Elaine C., Couch Gregory S., Croll Tristan I., Morris John H., and Ferrin Thomas E.. ”UCSF ChimeraX: Structure visualization for researchers, educators, and developers.” Protein Science 30, no. 1 (2021): 70–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Bell JM, Chen M, Durmaz T, Fluty AC, Ludtke SJ. New software tools in EMAN2 inspired by EMDatabank map challenge. J Struct Biol. 2018. Nov;204(2):283–290. doi: 10.1016/j.jsb.2018.09.002. Epub 2018 Sep 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Alnabati Eman, Terashi Genki, and Kihara Daisuke. ”Protein Structural Modeling for Electron Microscopy Maps Using VESPER and MAINMAST.” Current Protocols 2, no. 7 (2022): e494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Wriggers Willy. ”Using Situs for the integration of multi-resolution structures.” Biophysical reviews 2, no. 1 (2010): 21–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53] **. Bronstein Michael M., Bruna Joan, Cohen Taco, and Veličković Petar. ”Geometric deep learning: Grids, groups, graphs, geodesics, and gauges.” arXiv preprint arXiv:210413478. (2021). A (proto-) book on geometric deep learning about representational learning architectures and exploiting the symmetries of data therein.