Figure 1. Many enzymes from intermediary metabolism form nondirectional filaments with dihedral symmetry.
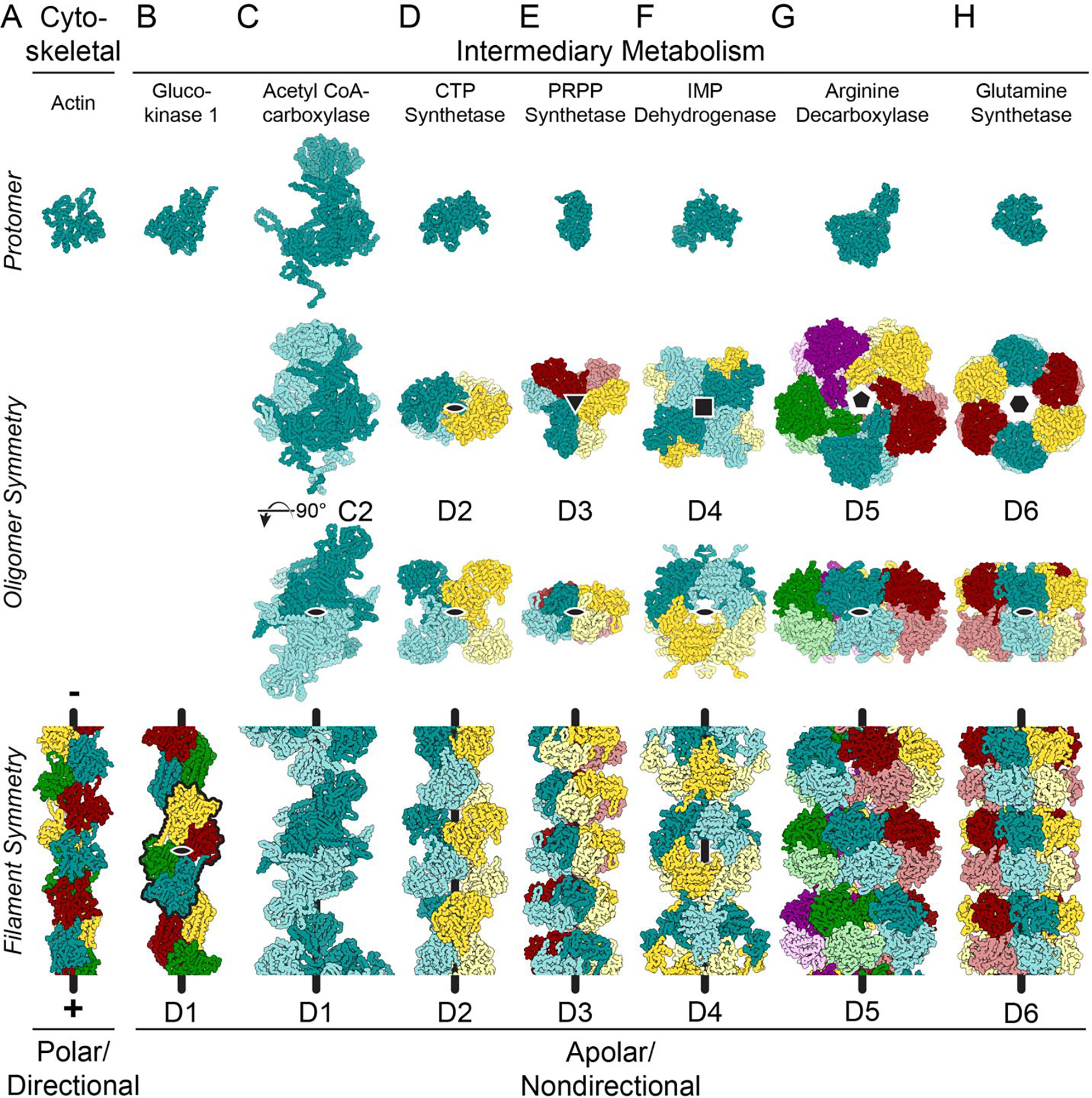
Images show mainchain C, N, and Cα atoms only. The symmetry for the helical axes is shown in the oligomer (black shapes, second row); one, two-fold axis is shown on the oligomer (black oval, third row); and the helical axis is shown as a black line on the filaments (bottom row). A. Cytoskeletal proteins, like actin (shown - PDB ID 6DJM [60]) and tubulin assemble from monomers or asymmetrical dimers, which produces a directional filament. The filaments grow and shrink from a defined pole of the filament structure, and the longitudinal interfaces within the filaments are comprised of residues found on opposite sides of the protein. B. While glucokinase 1 filaments (6PDT) assemble from monomers, the repeating units in the filament (black outline) assemble with C2 symmetry around an axis (black oval) perpendicular to the helical axis, generating a filament with overall D1 symmetry. C. Symmetric C2 dimers can also assemble into filaments with D1 symmetry (Acetyl CoA-carboxylase: 6G2D). The symmetry axis perpendicular to the helical axis is shown in both the oligomer and the filament (black oval). D-H. Enzymes found in intermediary metabolism have dihedral symmetry within the oligomer which is propagated in the filament form of the enzyme (CTP synthase: 6PK4; PRPP synthetase: 8DBF; IMP dehydrogenase: 7RGL/7RGM; arginine decarboxylase: 7PK6; glutamine synthetase: 7W85).
