Figure 3: Examples of different assembly-based mechanisms for tuning allosteric regulation.
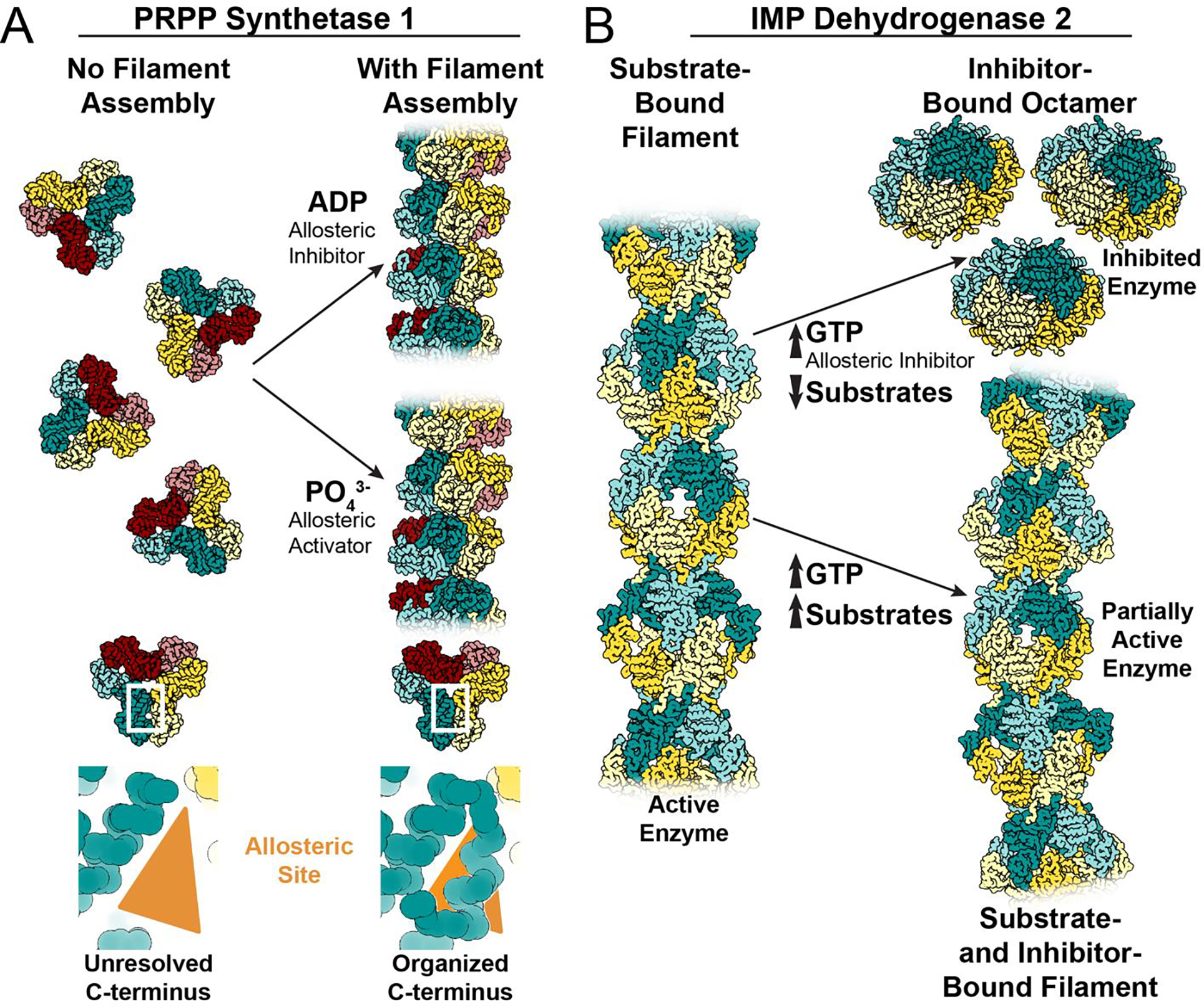
A. Without filament formation, the C-terminus of PRPP synthetase 1 is unresolved. Filament assembly organizes the amino acids in the C-terminus, stabilizing the allosteric site to increase affinity for effectors, increasing sensitivity to both positive and negative regulation (PDB IDs 8DBD and 8DBF). B. IMP dehydrogenase 2 assembles filaments in the active, extended conformation when bound to substrates (PDB ID 6U8E/6N8N). The allosteric inhibitor GTP stabilizes an inactive, compressed octamer conformation when substrate concentrations are limiting (PDB ID 6UAJ). When substrate and inhibitor concentrations are high, filament assembly contacts resist the GTP-induced compression to retain activity in the filament at GTP concentrations which completely inhibit free octamers (PDB ID 6U8R/6U8S). The partially active, bent filaments merge the competing signals that arise in an environment with high concentrations of both substrate and inhibitor.
