Figure 7. Variants in HARs interact to tune enhancer activity.
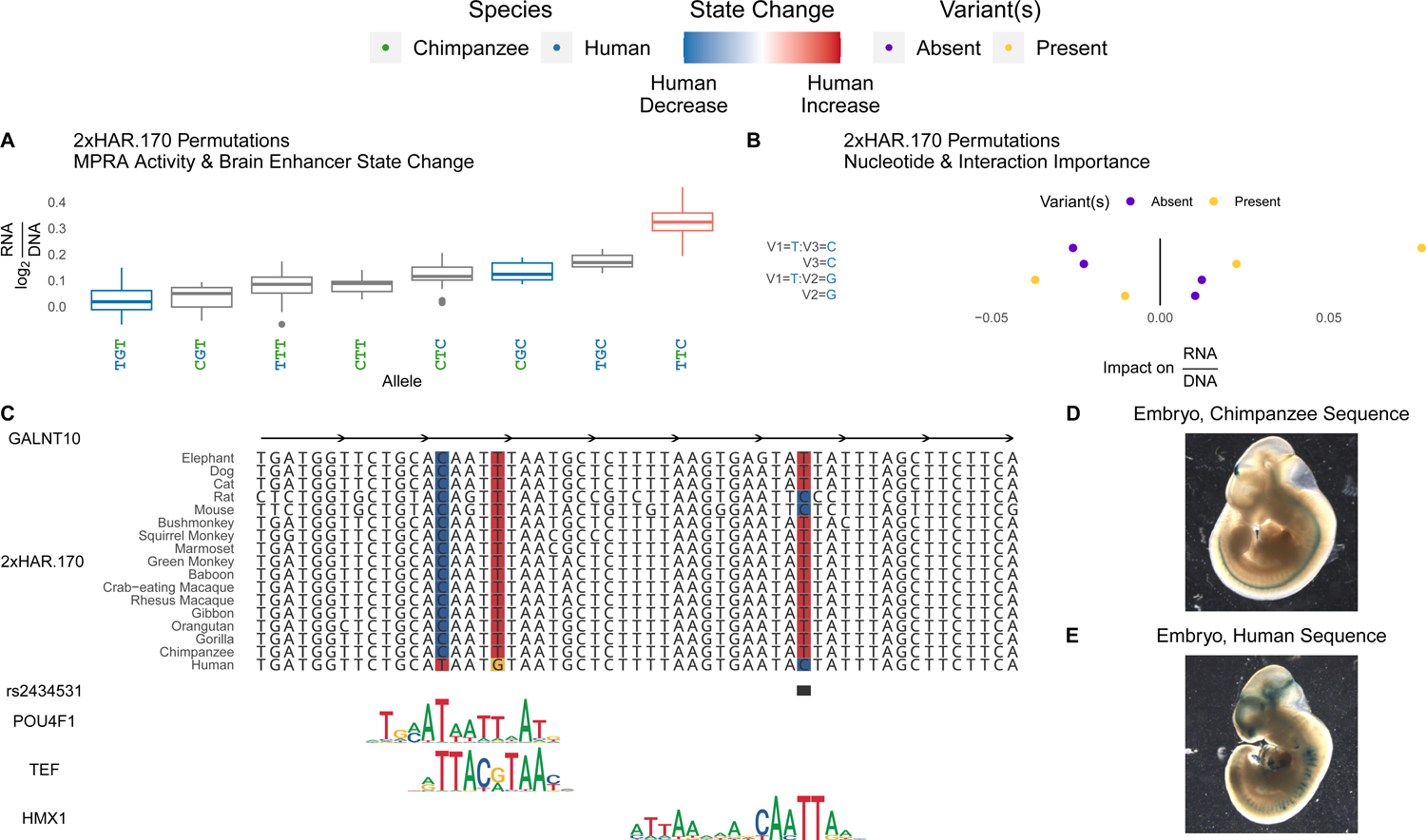
(A) All evolutionary intermediates between chimpanzee and human alleles of 2xHAR.170 were tested via lentiMPRA. Individual variants showed a range of effects on activity (y-axis; log2(RNA/DNA)) that correlated with Sei predicted effects on brain enhancer activity (red = human increase, blue = human decrease). Oligos containing multiple variants revealed interactions between variants that were untestable with Sei (no color). (B) We assessed the importance of each variant using a gradient boosting model that predicts lentiMPRA activity of each permutation using the presence or absence of the human allele at each of the three variants. Interactions between multiple variants (separated by colons on the y-axis) were included as predictors alongside main effects (no colon) to assess their predictive importance. This model confirmed the importance of specific variant interactions (x-axis, positive = higher predicted activity, negative = lower activity). Variant names consist of a V followed by the variant number (1, 2 or 3 ordered from 5’ to 3’), and the allele is shown after the equal sign. Present (yellow) indicates the expected change in enhancer activity for oligos that have the allele denoted on the y-axis, while absent (purple) shows the expected change for oligos that lack the allele. Yellow points at positive impact on RNA/DNA values means that variant or variant combination increases enhancer activity on average across oligos with other variants, while purple points at positive values mean the variant or variant combination decreases activity (i.e., activity is higher when absent). (C) 2xHAR.170 is a candidate intronic enhancer of GALNT10, acquiring a human-specific change from C to T that enhances POU4F1 and TEF binding in our footprint analysis. The human polymorphism rs2434531 is an eQTL for GALNT10, and we predict that the derived allele enhances binding of the repressor HMX1. The HMX1 and TEF footprints were detected in an independent brain footprinting study39. Both results are supported by Sei predictions (4E), lentiMPRA activity (A), and differential activity between the chimpanzee (D) and human (E) sequences in the forebrain and midbrain of transgenic mouse embryos. Adapted from6. The eQTL (rs2434531) is in linkage disequilibrium with a schizophrenia GWAS variant (rs11740474)51. In neuronal cells, 2xHAR.170 is bound by FOXP2, as well as other enhancer-associated proteins (ISL1, HAND2, PHOX2B, FOSL2) and chromatin modifiers (EZH2, SMARCA2, SMARCC1)60.
