Figure 2. The landscape of NS1 structures.
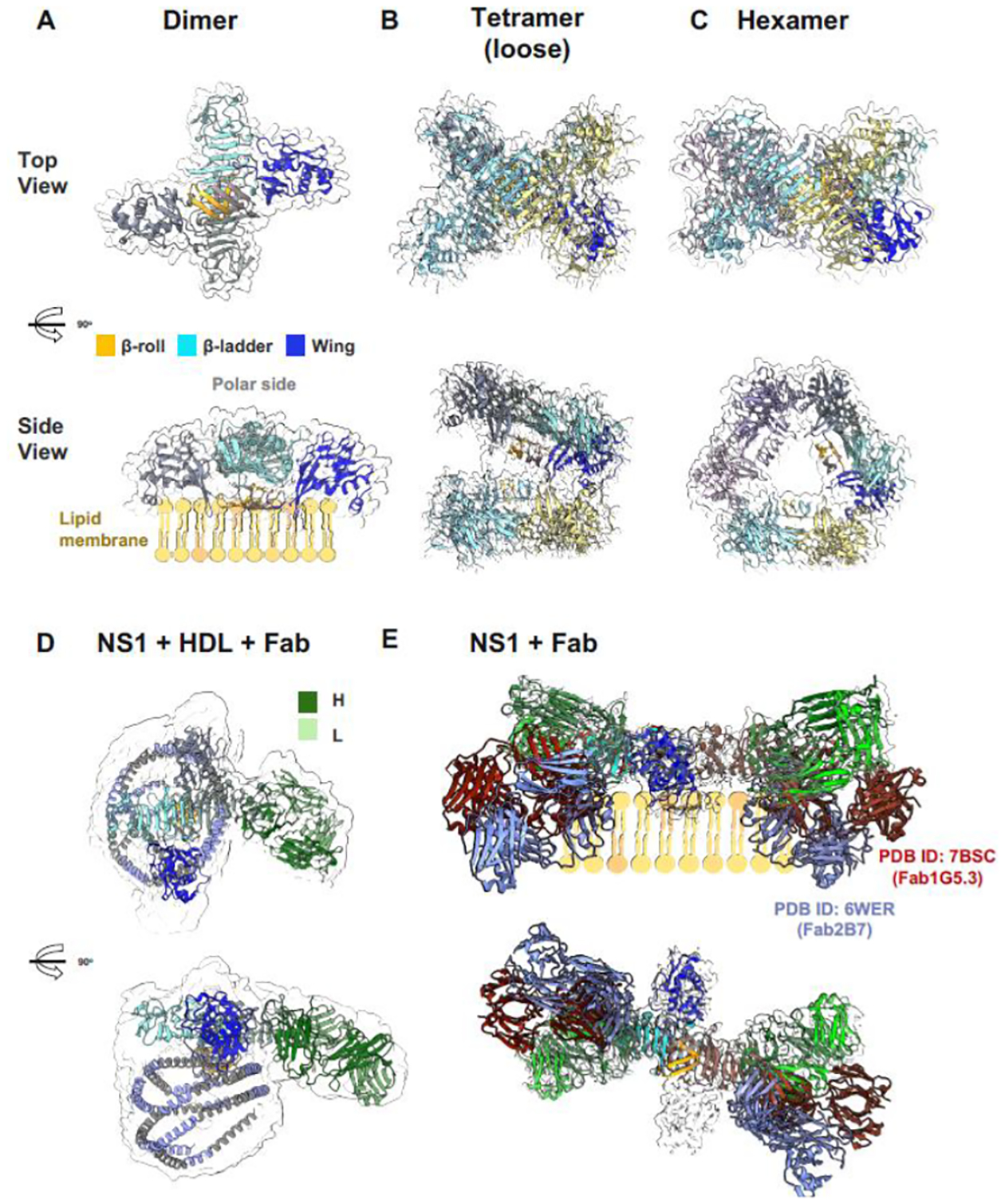
(A) Dimeric NS1 cartoon model in a simulated transparent map at 2.5 Å at a low contour of 0.2. One of the monomers is colored by its three-domain architecture, a hydrophobic β-roll (residues 1–29, orange), an a/b wing (residues 38–151, blue), and a rigid β-ladder (residues 181–352, cyan). The dimer has a distinct crossed shape with the wings extending from the central β-ladder which has an extended β-sheet that faces the β-roll as the membrane-associated surface (side view) and a “spaghetti loop” on the opposite polar outer face that lacks structured elements. (B) Tetrameric and (C) hexameric recombinant secreted NS1 (sNS1) forms as confirmed by Bo et al. (2021) cryoEM structures [1], modelled using the dimeric NS1 structure coupled with a simulated transparent map at 2.5 Å at the high contour of 1.0 and at 6.0 Å at the low contour of 0.3 respectively. The second NS1 dimer is coloured in yellow and blue for each monomer chain. The third NS1 dimer for the hexamer model is coloured in light and dark purple. (D) Immunoaffinity-purified sNS1 form from infected cell cultures as reported by Chew et al (2022) [2]. Model of sNS1wt: Fab56.2 predicted structures rigid body fitted in the CryoEM map (transparent grey, contoured at 0.14) with a correlation value of 0.75 to the fitted regions (map simulated from atoms at 5 Å). The antibody Fab56.2 domain is coloured in dark and light green for the Heavy (H) and Light (L) chains respectively. The predicted apoA-I, major protein component of HDL, model is coloured in intervals of grey and light purple representing its 11 and 22 residue alpha helical repeats. ApoA-I model is fitted in the spherical map region with its orientation informed by cross-linking mass spectrometry. (E) Comparison of the NS1 binding by anti-Denv Fab56.2 to earlier published NS1:Fab structures namely 2B7 (PDB ID: 6WER) [3] and 1G5.3 (PDB ID: 7BSC) overlayed on the lipid membrane cartoon for the side view and the top view shown at the bottom depicting the differing angles that they bind to the β-ladder. Figure prepared using ChimeraX.
