Figure 4. Predicted protein structure of NS4B and its inhibitors.
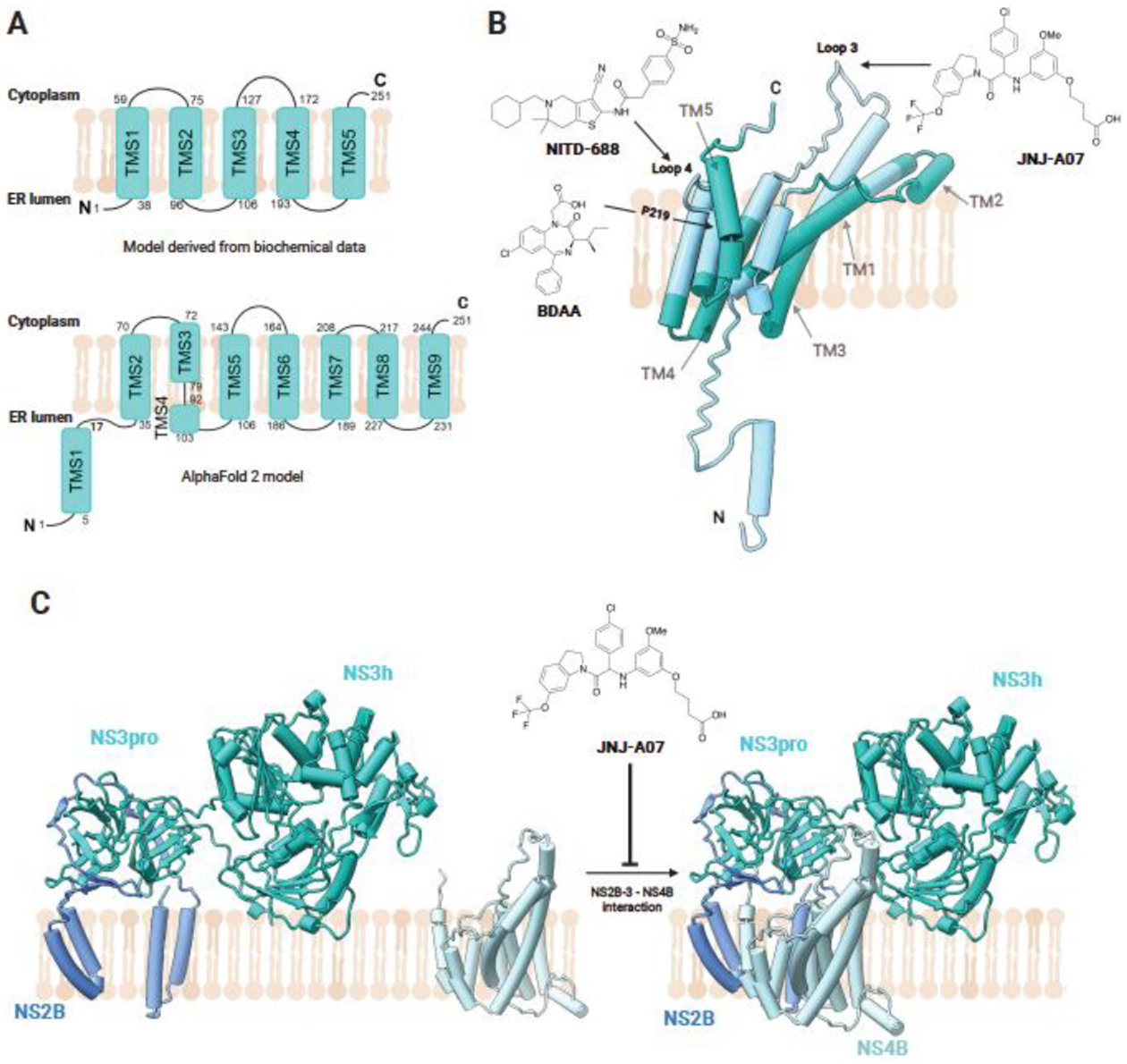
(A) Comparison between NS4B topology derived from biochemical data, adapted from Bhardwaj, et al., and topology of the AF2 model. (B) Schematic of AF2-predicted structural organisation of NS4B in the ER membrane, with proposed transmembrane domains based on biochemical studies labelled and colored in dark green and cytoplasmic facing regions coloured in cyan. Sites of interaction between NS4B and the compounds JNJ-A07, NITD-688, and BDAA are annotated. JNJ-A07 alters the conformation of loop 3 of NS4B, occurring between TM3 and TM4. BDAA, a small molecule inhibitor of YFV, binds P219 in TM5 of NS4B. NITD-688 has also been identified to bind to NS4B. (C) Effect of JNJ-A07 binding on the interaction of NS2B3-NS4B. JNJ-A07 blocks NS3-NS4B interaction and complex formation but does not disrupt existing NS3-NS4B complexes. Created with Biorender.
