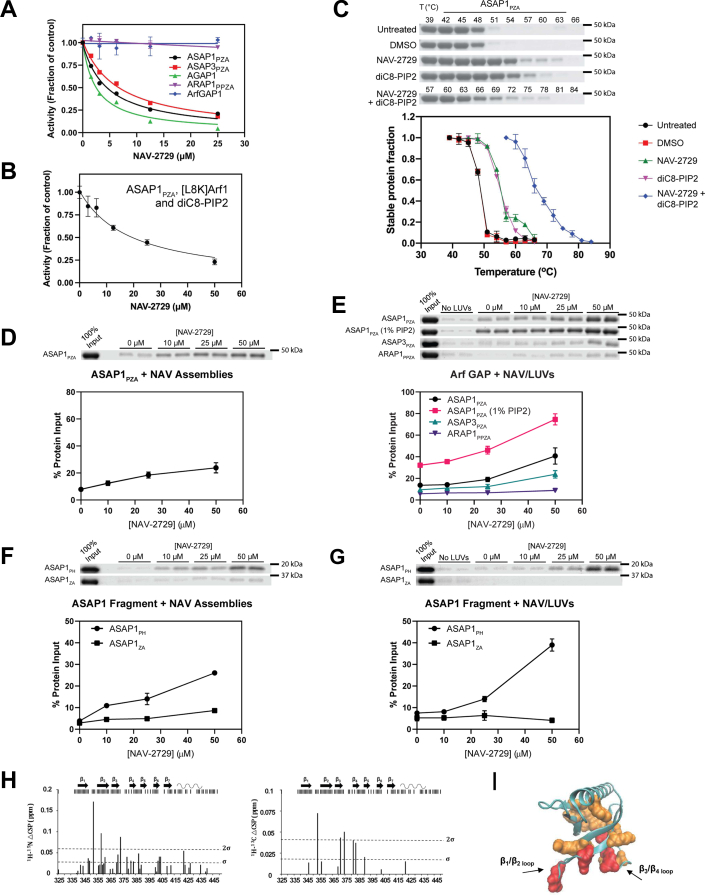
Figure 6.
NAV-2729 binds to and inhibits specific Arf GAPs.A, effect on GAP activity. NAV-2729 was titrated into reactions containing the indicated Arf GAPs, myrArf1•GTP, and LUVs as described in Experimental procedures. A representative experiment of 4 with ASAP1 and ASAP3, and 5 with AGAP1, are shown. B, NAV-2729 inhibits ASAP1 in the absence of a membrane. GAP activity was determined using ASAP1 as the GAP, [L8K]Arf1•GTP as the substrate and diC8-PIP2 as the activating ligand. Results are representative of six experiments. C, ASAP1 binds to NAV-2729 and diC8-PIP2. Thermal shift assays were conducted as described in Figure 4A. Representative gels and the summary of six experiments are shown. D, ASAP1 binds NAV-2729 supramolecular assemblies. ASAP1 was tested for binding as described in Figure 4B. Representative gel and summary of four experiments are shown. E, ASAP1 and ASAP3, but not ARAP1, bind to LUVs containing NAV-2729. These assays were the same as described in Figure 4C. In addition to the 0% PIP2 condition, ASAP1 was tested with LUVs containing 1% PIP2. Summaries of ≥3 experiments are shown. F, the PH domain, but not Arf GAP or ankyrin repeats (ZA construct) of ASAP1, binds to NAV-2729 supramolecular assemblies. Binding was determined as described in Figure 4B. Summary of three experiments is shown. G, the PH domain, but not Arf GAP or ankyrin repeats (ZA construct) of ASAP1, binds to NAV-2729 in LUVs. These assays were the same as described in Figure 4C. Three experiments are summarized. H, interaction between NAV-2729 and ASAP1PH at the membrane interface. The top of each chart shows a summary of data collected. Black bars correspond to residues where data could be collected. Most of the missing residues come from exposed HN exchanging with the solvent at pH 7.4 (unstructured N-terminal stretch between residues 325 and 339 and loops). Charts show chemical shift perturbation differences (△CSP) between 1H-15N CS (Left) or 1H-13C CS (Right) for ASAP1 PH bound to nanodiscs with NAV-2729 and nanodiscs without NAV-2729. Nanodiscs with NAV-2729 contained four NAV-2729 molecules per nanodisc. I, △CSPs are mapped on the crystal structure of the ASAP1 PH domain (PDB 5C79) (43). Residues colored in red have △CSP larger than 2σ and residues colored in orange have △CSP larger than σ, where σ is the SD calculated over all △CSPs. Arf, ADP-ribosylation factor; CSP, chemical shift perturbation; GAP, GTPase-activating protein; LUV, large unilamellar vesicle; PH, pleckstrin homology; PIP2, phosphatidylinositol 4,5-bisphosphate.