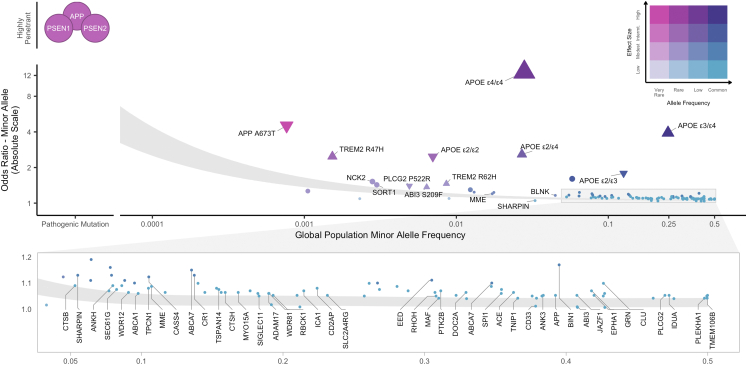
Fig. 1.
Genetic architecture of AD/dementia highlighting ADAD mutations and 81 genome-wide significant loci. Genetic variants associated with disease are often conceptualized along two dimensions–variant effect size and population minor allele frequency. Highly penetrant mutations in APP, PSEN1, PSEN2 that segregate with autosomal dominant AD are extremely rare and have large effect sizes. Variants discovered by genome-wide associations are mostly common to low-frequency with small effect sizes. To date, AD/dementia GWAS have identified 101 independent AD-associated single nucleotide polymorphisms across 81 genome-wide significant (p < 5e-8) loci. The shaded area illustrates 80% power to detect genome-wide significant association for variants at a given effect size and population frequency between an effective sample size of 382,472 from Bellenguez et al. (2022) (top) and 1 million (bottom), assuming 0.18 AD prevalence. Odds ratios are reported on the absolute scale, with triangles indicating directionality for APOE genotypes and rare variants with moderate-high SnpEff impact annotations. Effect sizes for APOE genotypes and APP Ala673Thr were obtained from Reiman et al.1 and Jonsson et al.,16 respectively. Labeled loci indicate candidate causal genes prioritized by Bellenguez et al. (2022). Variant MAFs and APOE genotype frequencies were obtained from the gnomAD global population (GRCh37 v2.1.1).