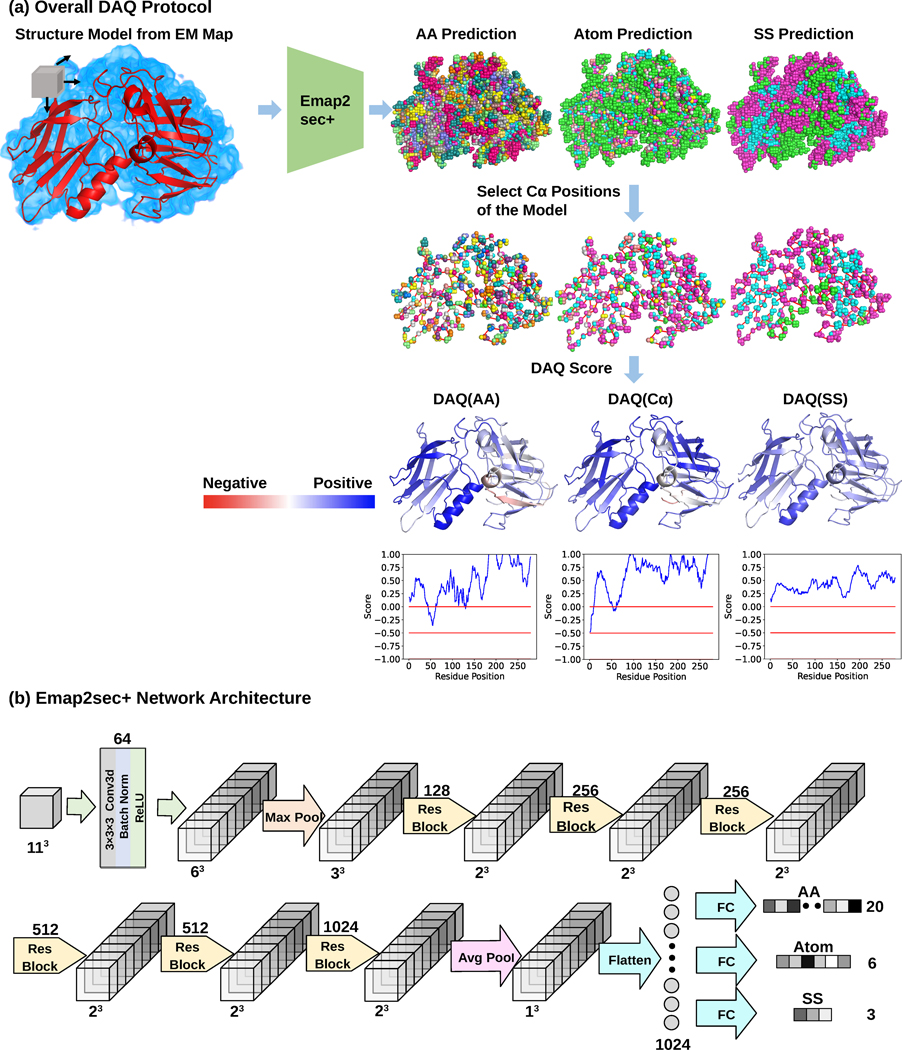
Fig. 1. Overview of DAQ.
DAQ is a residue-wise local quality estimation for protein models from cryo-EM maps based on upgraded Emap2sec+. The example used here is the rNLRP1-rDPP9 complex (PDB-ID: 7CRW, chain A) and the EM map from which the structure was built (EMD-30458). a. DAQ protocol. Emap2sec+ scans an EM map with a box of a 11*11*11 Å3 size with a stride of 1 Å and outputs the probabilities of amino acid type, atom type, and secondary structure type for the center position of the box. Next, the probabilities at Cα positions of the structure model are gathered. Then, DAQ(AA), DAQ(Cα), and DAQ(SS) are further calculated as log-odds scores using the average probability for the corresponding property across the entire model. In this figure, higher values (blue) indicate higher quality indicated by DAQ, while red indicate lower quality of the local structures by DAQ. b. a detailed network architecture of upgraded Emap2sec+, which were used to compute the probability values. It has 6 residual blocks and outputs the amino acid, atom, and secondary structure probability for an input box.