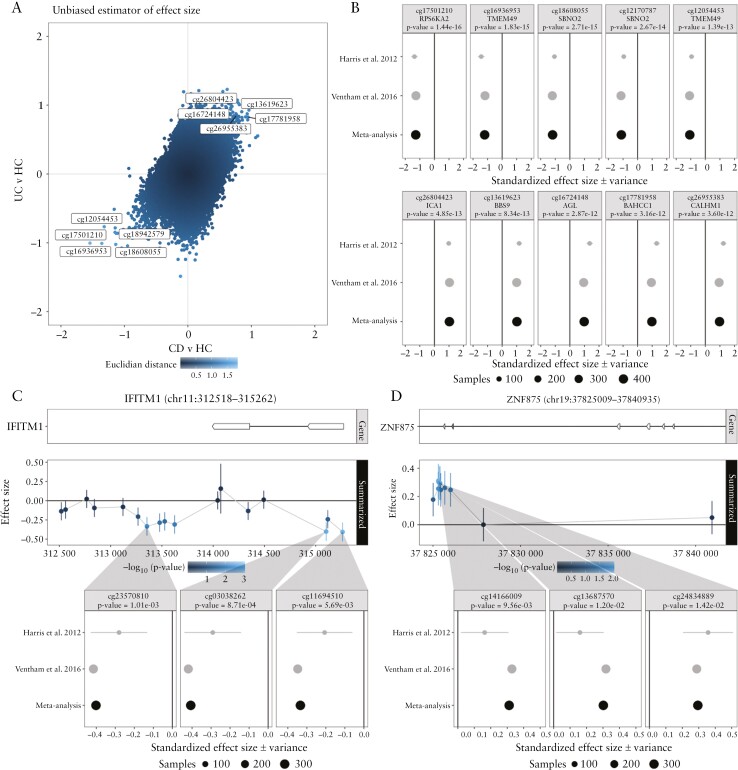
Figure 4.
Meta-analysis of the comparisons Crohn’s disease [CD] with ulcerative colitis [UC] and inflammatory bowel disease [IBD] with healthy controls [HC] using data from Harris et al. 2012 and Ventham et al. 2016. [A] A scatter plot of the included CpGs representing aggregated effect size of CD with HC and UC with HC on the x- and y-axis, respectively. Colour intensity is proportional to the distance of each CpG to the origin [0,0]. Annotated are the CpGs that are concordantly hyper- and hypomethylated in both CD and UC, and hence IBD, relative to HC. [B] Forest plots representing the standardized effect size [SES] of interest per study of the five most hyper- and hypomethylated IBD-associated DMPs annotated with the associated gene and p-value from the meta-analysis. Genomic visualization and forest plots of the estimated difference in methylation for the CD/UC discordant genes [C] IFITM1 and [D] ZNF875, alongside for the most significant differentially methylated probes.