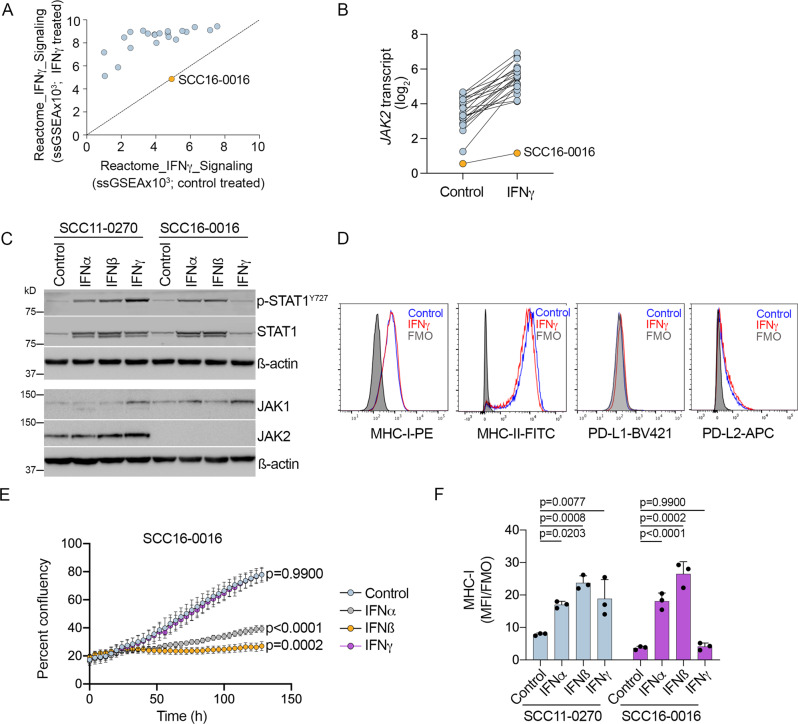
Fig. 1. IFNγ response in PD1 PROG cell lines.
A ssGSEA scores for the Reactome_Interferon_Gamma_Signaling gene set in 22 PD1 PROG melanoma cell lines treated with BSA control (x-axis) or 1000 U/ml IFNγ (y-axis) for 24 h. The SCC16-0016 PD1 PROG did not respond to IFNγ stimulation and is highlighted. B JAK2 transcript expression in matched BSA control and IFNγ-treated PD1 PROG cell lines (n = 22). C Accumulation of phosphorylated STAT1 (p-STAT1Y727), STAT1, JAK1, and JAK2 in the SCC11-0270 (responded to IFNγ) and SCC16-0016 (did not respond to IFNγ) PD1 PROG cell lines treated with BSA control, IFNα, IFNβ, or IFNγ (all at 1000 U/ml) for 24 h. D Representative histograms showing levels of MHC-I, MHC-II, PD-L1, and PD-L2 expression in SCC16-0016 cell line after BSA control (blue) or 1000 U/ml IFNγ (red) treatment for 72 h. Fluorescence minus one (FMO) controls are shown as shaded histograms. E Representative proliferation curve of SCC16-0016 cells, measured as percent confluence every 4 h, for up to 120 h, after treatment with BSA control, IFNα, IFNβ, or IFNγ (all at 1000 U/ml). Data shown are mean ± sd (six images per treatment per time point). Area under the curve data of biological triplicates were compared using one-way ANOVA with Dunnett correction for multiple comparisons, and adjusted p-values are shown. F Cell surface expression (median fluorescence intensity stained divided by fluorescence minus one control, MFI/FMO) of MHC-I in SCC11-0270 and SCC16-0016 PD1 PROG cell lines treated with BSA control, IFNα, IFNβ, or IFNγ (all at 1000 U/ml) for 72 h. Data shown are mean ± sd (n = 3 biologically independent experiments) and were compared using one-way ANOVA with Dunnett correction for multiple comparisons, and adjusted p-values are shown. Source data are provided as a Source Data file.