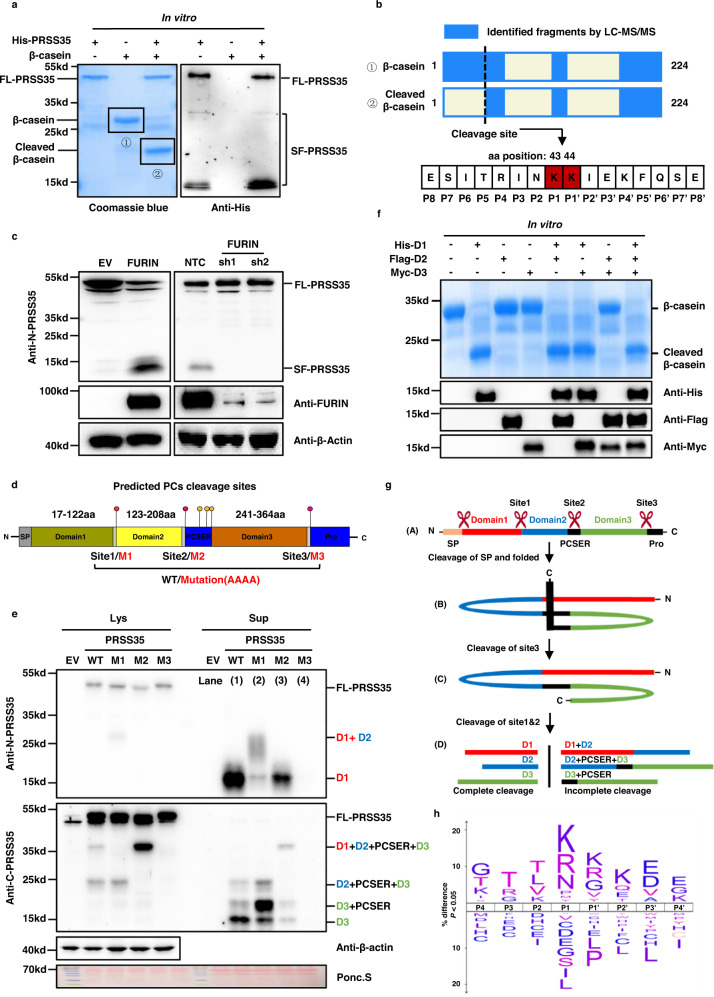
Fig. 2. PRSS35 activated by FURIN functions as a protease.
a His-PRSS35 protein purified from E. coli was incubated with β-casein protein at 37 °C overnight, followed by SDS-PAGE and coomassie brilliant blue staining (left panel). PRSS35 signal was determined by western blot with anti-His antibody (right panel). FL full length, SF short form. b Bands from (a) were analyzed by mass spectrometry to identify PRSS35 cleavage sites. Schematic of β-casein coverage by LC–MS/MS identified peptides and schematic of PRSS35 cleavage site in β-casein (upper panel). PRSS35 cleavage of β-casein at K43-K44 was monitored by LC–MS/MS (lower panel). c Western blot analysis of PRSS35 protein levels in HepG2 cells stably expressing FURIN or EV (left panel) or in HepG2 cells stably expressing shFURIN or NTC (right panel). β-actin served as loading control. FL full length, SF short form. d Schematic diagram of the PRSS35 protein and the predicted proprotein convertases (PCs) cleavage sites in PRSS35. Conserved sequences of proprotein convertases cleavage sites in PRSS35 and corresponding mutant sequences are shown. PCSER: PCs cleavage sites enriched region. e Western blot analysis of intracellular and extracellular PRSS35 protein levels with anti-N-PRSS35 and anti-C-PRSS35 antibodies using PLC cells expressing PRSS35-WT, PRSS35-M1, PRSS35-M2, PRSS35-M3 or EV. Lys lysate (intracellular proteins), Sup supernatant (extracellular proteins), FL full length, PCSER PCs cleavage sites enriched region. D1: PRSS35 domain1. D2: PRSS35 domain2. D3: PRSS35 domain3. f E. coli purified His-D1, Flag-D2 and Myc-D3 protein alone, or in combination, were incubated with β-casein protein at 37 °C overnight, followed by SDS-PAGE and coomassie brilliant blue staining. His-D1, Flag-D2 and Myc-D3 signals were determined by western blot with anti-His, anti-Flag and anti-Myc antibodies. FL full length, SF short form, D1 PRSS35 domain1, D2 PRSS35 domain2, D3 PRSS35 domain3. g The working model depicts the sequential self-cleavage of PRSS35 protein. h An iceLogo (upper panel) generated from 1318 purified human FL-PRSS35 and PRSS35-domain1 cleavage sites identified from PLC, HepG2, Hep3B and 293T cells with the human Swiss-Prot proteome as reference set (lower panel). In this logo, PRSS35 cleavage occurred at the peptide bond between residues position1 (P1) and position1′ (P1′). The blotting experiments were repeated at least three times with biological replicates (a, c, f, e). Source data are provided as a Source Data file.