Abstract
Central Nervous System (CNS) embryonal tumors represent a heterogeneous group of highly aggressive tumors occurring preferentially in children but also described in adolescents and adults. In 2021, the CNS World Health Organization (WHO) classification drastically changed the diagnosis of the other CNS embryonal tumors including new histo-molecular tumor types. Here, we report a pediatric case of a novel tumor type among the other CNS embryonal tumors classified within the methylation class “CNS Embryonal Tumor with BRD4–LEUTX Fusion”. The patient was a 4-year girl with no previous history of disease. For a few weeks, she suffered from headaches, vomiting and mild fever associated with increasing asthenia and loss of weight leading to a global deterioration of health. MRI brain examination revealed a large, grossly well-circumscribed tumoral mass lesion located in the left parietal lobe, contralateral hydrocephalus and midline shift. Microscopic examination showed a highly cellular tumor with a polymorphic aspect. The majority of the tumor harbored neuroectodermal features composed of small cells with scant cytoplasm and hyperchromatic nuclei associated with small “medulloblastoma-like” cells characterized by syncytial arrangement and focally a streaming pattern. Tumor cells were diffusely positive for Synaptophysin, CD56, INI1 and SMARCA4 associated with negativity for GFAP, OLIG-2, EMA, BCOR, LIN28A and MIC-2. Additional IHC features included p53 protein expression in more than 10% of the tumor’s cells and very interestingly, loss of H3K27me3 expression. The Heidelberg DNA-methylation classifier classified this case as “CNS Embryonal Tumor with BRD4:LEUTX Fusion”. RNA-sequencing analyses confirmed the BRD4 (exon 13)–LEUTX (exon 2) fusion with no other molecular alterations found by DNA sequencing. Our case report confirmed that a new subgroup of CNS embryonal tumor with high aggressive potential, loss of H3K27me3 protein expression, BRDA4–LEUTX fusion, named “Embryonal CNS tumor with BRD4–LEUTX fusion”, has to be considered into the new CNS WHO classification.
Keywords: Brain tumor, CNS embryonal tumor, DNA methylation, H3K27me3 protein expression, BRD4, LEUTX
Introduction
Central Nervous System (CNS) embryonal tumors represent a heterogeneous group of highly aggressive tumors occurring preferentially in children but also described in adolescents and adults [17]. Among them, the diagnosis of medulloblastomas using integration of histopathological and molecular features is exemplified [18]. Nevertheless, for the other CNS embryonal tumors, we are at the first step of the emergence of this histo-molecular diagnostic approach. This has been underlined by Sturm et al. who revealed in 2016 new molecular entities among these poorly differentiated embryonal tumors [17]. In 2021, the CNS World Health Organization (WHO) classification drastically changed the diagnosis of the other CNS embryonal tumors including new histo-molecular tumor types: CNS neuroblastoma, FOXR2-activated, CNS tumor with BCOR internal tandem duplication (ITD) and cribriform neuroepithelial tumor [12]. These subtypes are defined by essential criteria combining histopathological, immunohistochemical, molecular as well as epigenetics data [12]. This underlined the heterogeneity of these tumors and the need to further characterization. Here, we report a pediatric case of a novel tumor type among the other CNS embryonal tumors classified within the methylation class (MC) “CNS Embryonal Tumor with BRD4:LEUTX Fusion” (calibrated score (cs): 0.99) which is not yet integrated into the CNS WHO classification.
Case presentation
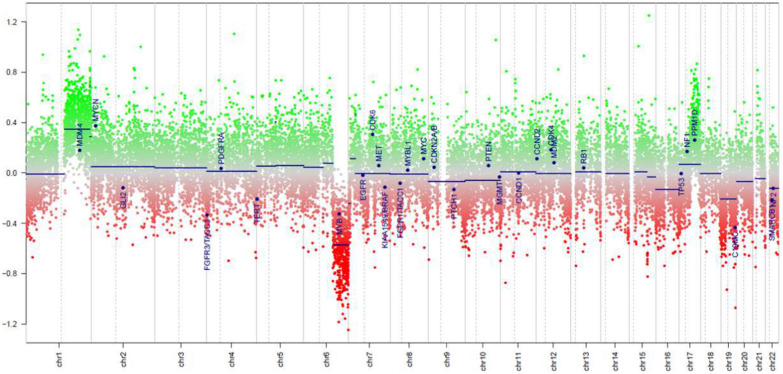
The patient was a 4-year girl with no previous history of disease except for intrauterine growth restriction with a birth weight of 1900 g. For a few weeks, she suffered from headaches, vomiting and mild fever associated with increasing asthenia and loss of weight leading to a global deterioration of health. An ophthalmological examination revealed papilledema. A Computed Tomography (CT)-Scan and subsequently a Magnetic Resonance Imaging (MRI) of the brain were therefore obtained, demonstrating a large, grossly well-circumscribed tumoral mass lesion located in the left parietal lobe, contralateral hydrocephalus and midline shift (Fig. 1). There were signs of subependymal and leptomeningeal dissemination, along with several smaller size tumoral lesions both supra- and infratentorial. The proposed radiological diagnosis was that of a metastatic Atypical Teratoid Rhabdoid Tumor (ATRT). Intra-operative aspect confirmed the heterogeneity of the tumor, highly hemorrhagic, with numerous calcifications and necrosis. Microscopic examination showed a highly cellular tumor with a polymorphic aspect (Fig. 2). The majority of the tumor harbored neuroectodermal features composed of small cells with scant cytoplasm and hyperchromatic nuclei associated with small “medulloblastoma-like” cells characterized by syncytial arrangement and focally a streaming pattern. No Homer Wright or Flexner–Wintersteiner rosettes were reported. In some areas the cells harbored moderately bi-, multi-nucleated and pleomorphic nuclei with numerous “bizarre cells”. The mitotic activity was high with 5–10 mitoses per 2.3 mm2 and KI-67 proliferative index nearing 50%. Numerous apoptotic bodies were observed. Intratumoral desmoplasia was a prominent feature of this tumor as confirmed by Reticulin stain. Microvascular proliferation, extensive geographical necrosis, hemorrhage and calcifications were observed. The tumor was characterized by the lack of ganglion cells, ependymal rosettes, peri-vascular pseudo-rosettes, multilayered structures, nodular formation, cribriform structure, mesenchymal or epithelial differentiation, rhabdoid cells. Immunohistochemistry (IHC) analyses are illustrated in Fig. 3. Tumor cells were diffusely positive for Synaptophysin, CD56, INI1, SMARCA4 and ATRX associated with negativity for GFAP, OLIG-2, EMA, BCOR, LIN28A, EZHIP, MIC-2, Cytokeratin’s, SMA, Desmin and Myogenin. The combination of the morphological and IHC profiles led us to classify this case under “other CNS embryonal tumors, NOS”. Additional IHC features included p53 protein expression in more than 10% of the tumor’s cells and very interestingly, loss of H3K27me3 expression. Regarding molecular analyses, NGS sequencing revealed no mutations in genes commonly involved in CNS tumors such as ACVR1, IDH1/2, BRAF, H3F3A, HISTH3B/C, TP53, ATRX, TERT, SMARCB1 and EGFR genes, extensively described in [5, 11]. NGS testing suggested atypical Chromosome 1q profile with no loss or gain of heterozygoty of chromosome 19. Finally, using the Heidelberg DNA-methylation classifier [3], the case was classified as “CNS Embryonal Tumor with BRD4:LEUTX Fusion” (with a calibrated score (cs) of 0.99). Copy Number Variation (CNV) profile, provided by the online platform https://www.molecularneuropathology.org, suggested gain of chromosome 1q and loss of chromosome 6q (Fig. 4). We confirmed this diagnosis by RNA-sequencing analyses using kit FusionPlex Pan Solid Tumor v.2 (Archer Dx) which revealed BRD4–LEUTX fusion. The breakpoint locations are chr19:15355042 and chr19:40275167 for BRD4 (exon 13; NM_058243.2) and LEUTX (exon 2; NM_0001143832.1), respectively. Three weeks after sub-total surgical resection of the left parietal tumor, the residual tumor as well as secondary tumoral locations previously described doubled in size (Fig. 1). Numerous other lesions appeared in supratentorial and infratentorial regions and meningeal dissemination was observed in the spinal cord. Induction chemotherapy with VP16-Carboplatin according to PNET HR + 5 protocol (NCT00936156) was proposed but not feasible due to severe global deterioration. One month after the initial diagnosis, the patient passed away from neurological and clinical complications.
Fig. 1.
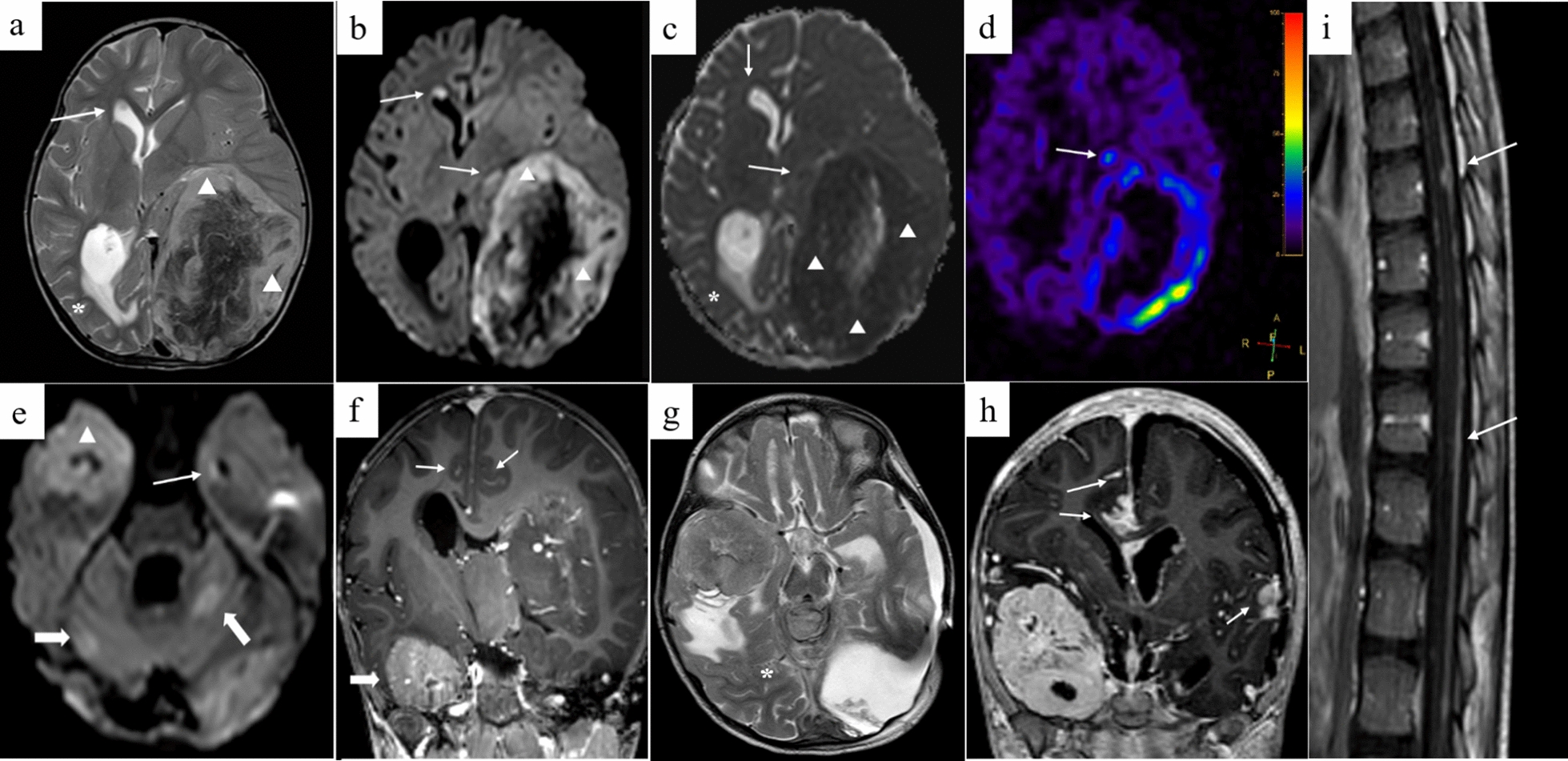
Radiological features. Pre-operative brain MRI a–f Images revealed a grossly well-marginated tumor mass lesion of approximately 8 × 9 cm located in the left parietal lobe. The lesion exerted severe mass effect on the ipsilateral lateral ventricle and adjacent brain parenchyma, resulting in contralateral active hydrocephalus (*in a and c) and midline shift. There was only mild peritumoral vasogenic edema. Signal intensity was strikingly heterogenous on all sequences suggesting the presence of core calcifications, necrosis and hemorrhage. The peripheral solid tumoral components displayed significant T2 prolongation (△ in a) and restricted diffusion on DW-images b, e and corresponding ADC map (△ in c), indicating high cellularity. ASL perfusion d demonstrated markedly increased CBF. Smaller size metastatic intraventricular (arrows in a–e), leptomeningeal (arrow in f) and parenchymal (bold arrows in e) lesions were present both supra-and infra-tentorially. The left parietal lobar lesion displayed faint, heterogenous contrast enhancement, whereas the right temporal lobe lesion enhanced avidly. The metastatic intraventricular deposits did not show any contrast uptake. Follow-up brain (g, h) and spinal cord (i) MRI: post contrast coronal T1-weighted (h) and sagittal T1-weighted (i) images of the brain and spinal cord, four weeks after diagnosis, demonstrated marked interval tumoral increase in size and progression of subependymal and leptomeningeal dissemination (arrows). Surrounding vasogenic edema was also increased (*in g). DW: diffusion-weighted; ADC, apparent diffusion coefficient; ASL, arterial spin labeling; CBF, cerebral blood flow
Fig. 2.
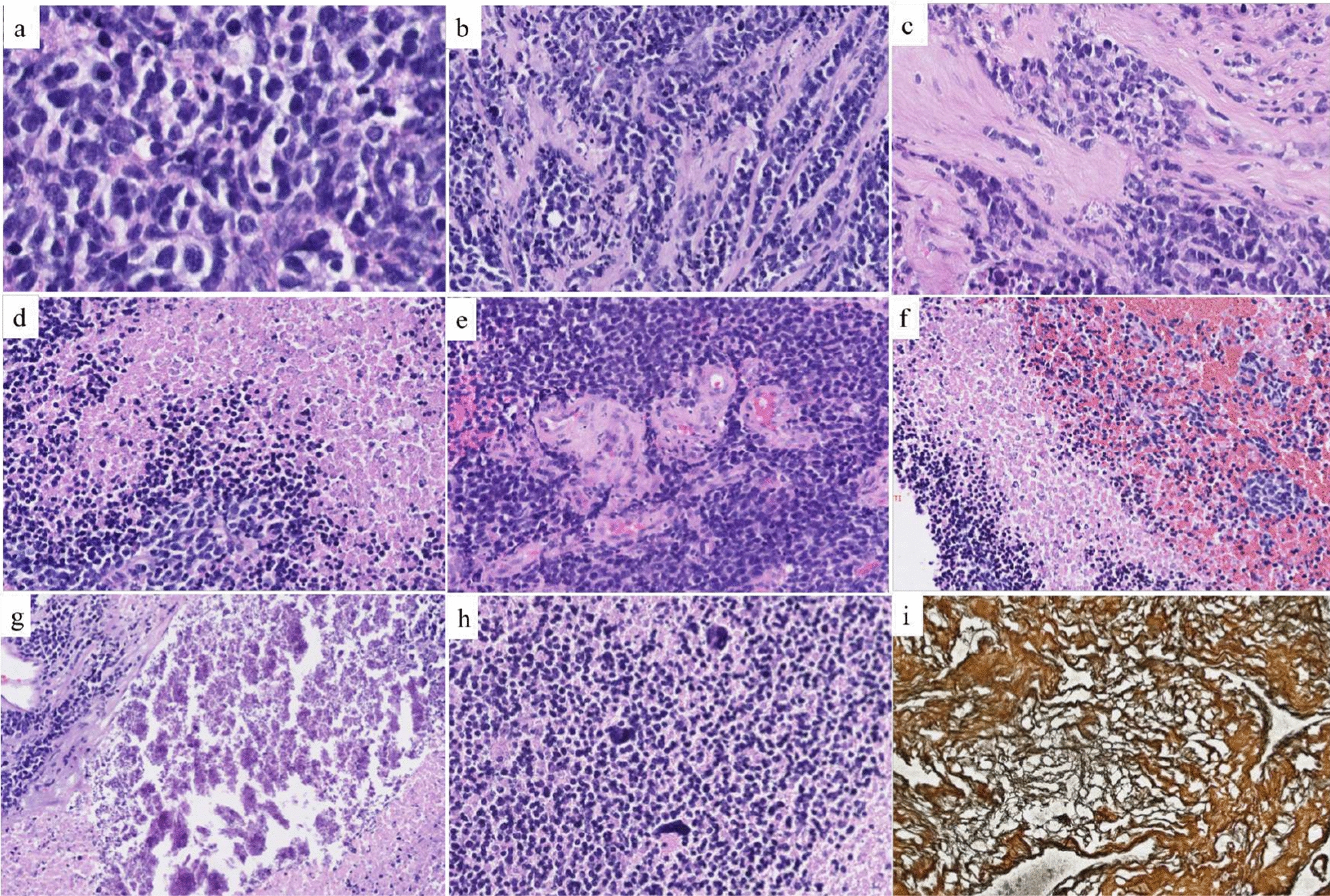
Histopathological features. a The tumor was highly cellular, composed of small cells with scant cytoplasm and hyperchromatic nuclei (HE, magnification 400×). b, c Intratumoral desmoplasia with focally tumoral cells disposed in a streaming pattern (HE, magnification 200×). d Tumoral geographical necrosis (HE, magnification 100×). e: Microvascular endothelial proliferation (HE, magnification 100×). f Intratumoral hemorrhage (HE, magnification 100×). g Calcifications (HE, magnification 200×). h Presence of numerous “bizarre cells” with high nuclear pleomorphism (HE, magnification 200×). i Reticulin stain highlighted intense desmoplasia (HE, magnification 200×). HE, Hematoxylin–eosin
Fig. 3.
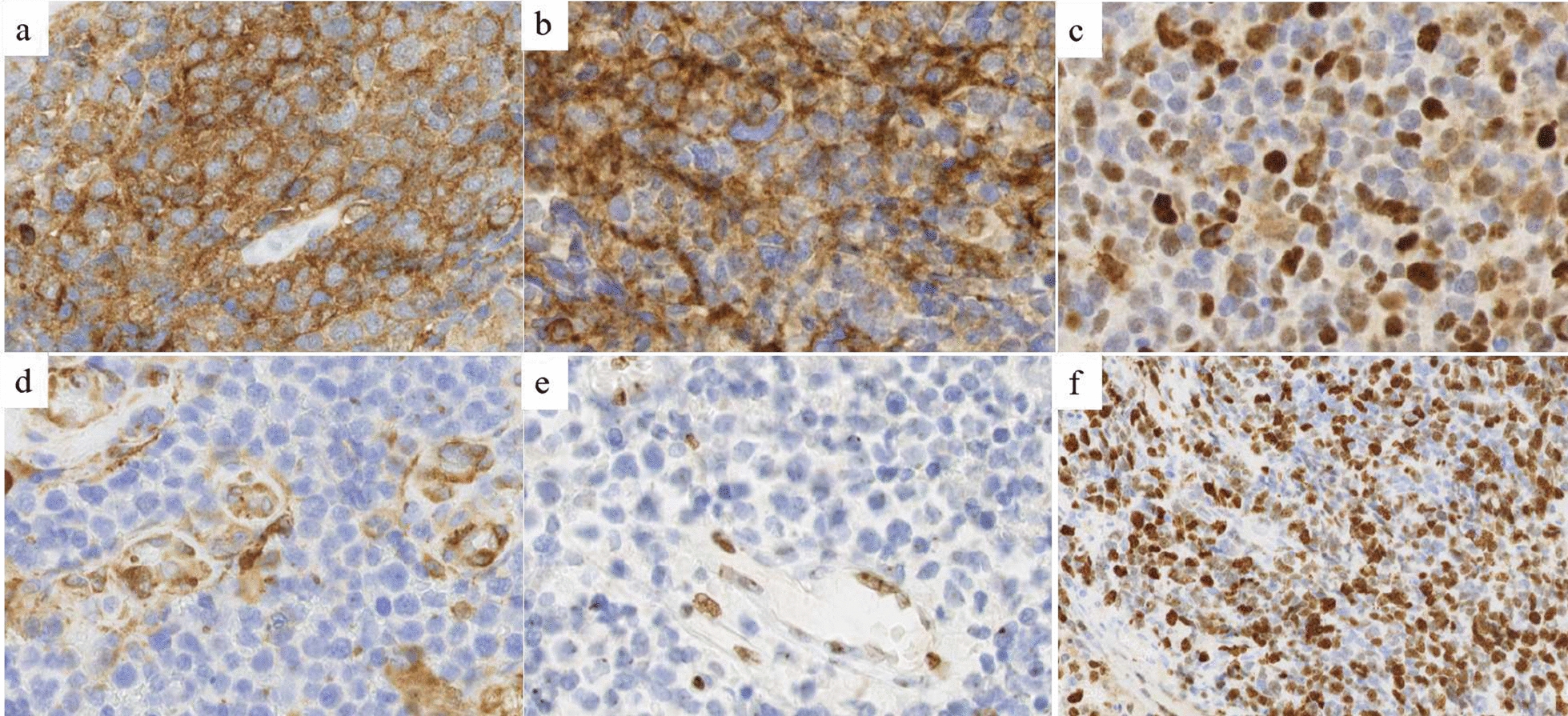
Immunohistochemical features. a Diffuse Synaptophysin immunopositivity (magnification 400×). b Diffuse CD56 immunopositivity (magnification 400×). c Intense nuclear immunopositivity of p53 protein in more than 10% of the tumoral cells (magnification 400×). d Loss of Filamine-A expression in tumoral cells with immunopositivity in endothelial cells (magnification 400×). e Loss of H3K27me3 expression in tumoral cells with immunopositivity in endothelial cells (magnification 400×). f High KI-67 proliferation index with some hotspot of more than 50% of immunopositivity in tumoral cells (magnification 200×)
Fig. 4.
Copy Number Variation (CNV) profile. DNA methylation profiling analyses by the Heidelberg Classifier V12.5 provided CNV profile of the case revealing gain of chromosome 1p and loss of chromosome 6q, provided by https://www.molecularneuropathology.org
Discussion and conclusion
BRD4–LEUTX fusion is a rare fusion which has only been described in the literature in one infant CNS embryonal tumor (BRD4 exon 11-LEUTX exon 2 fusion) in a comprehensive molecular profiling study of 252 high-risk pediatric cancers [19]. Outside the CNS, this fusion has been reported in one pediatric epithelioid malignant peripheral nerve sheath tumor and an alveolar rhabdomyosarcoma [2, 4]. Even though they share the same molecular alteration, the tumor locations, morphologies and IHC profiles were drastically different between our case and the BRD4–LEUTX pediatric sarcoma described by Barresi et al. [2]. Other LEUTX gene fusions including CIC gene (CIC–LEUTX fusion) were already reported in very few cases including one CNS embryonal tumor and an anaplastic pleomorphic astrocytoma [8, 16]. In these studies, no clinico-radiological, histological and DNA methylation profiling were reported. Interestingly, regarding CIC gene fusions, CIC–NUTM1 has been reported in CNS Ewing Sarcoma and represent the majority of CNS CIC-rearranged sarcomas, according to the CNS WHO classification [12, 16]. In 70–80% of cases, NUTM1 gene is involved in a balanced translocation with BRD4 gene in NUT carcinoma, which is a poorly differentiated carcinoma occurring mostly in children and adolescents [15]. Therefore, the tumors harboring BRD4–LEUTX, CIC–LEUTX, BRD4–NUTM1 and CIC–NUTM1 fusions seem to be mostly observed in children and young adults. Recent papers suggest that the bromodomain and extraterminal (BET) protein BRD4 could play an important role in the development of brain cancer. BRD4 protein is a chromatin reader protein involved in initial recognition of acetylated histones and plays a key role in epigenetic memory across cell divisions and transcription regulation [13, 20]. LEUTX homeobox protein is normally expressed during early embryogenesis and regulates genes associated with pluripotency and differentiation [10, 19]. Acetylated histones are bound to BRD4 to activate transcription. Therefore, BET bromodomain inhibitors blocking the bind between BRD4 and acetylated histones lead to transcriptional inactivation and have been reported to be promising therapeutic strategy in medulloblastomas and glioblastomas [7, 9]. The potential interchangeable ability of BRD4 and CIC genes regarding fusions with LEUTX and NUTM1 genes could be explained by the fact that both genes i.e. CIC and BRD4 are involved in the transcription regulation and associated with the recruitment of chromatin modifiers such as histone acetyl transferases [16]. Interestingly, in our case, loss of H3K27me3 expression was observed. While widely studied in neuroglial tumors leading to specific entities, the decrease of H3K27me3 expression is less described in CNS embryonal tumors with scarce data available regarding histone modifications alterations in these tumors. Interestingly, loss of H3K27me3 has been described by Alexandrescu et al. in primary intracranial sarcoma, DICER1-mutant [1, 14]. Studies reporting effects of targeting therapeutic EZH2 and BET inhibitors in ATRT [9, 21] lead us to propose the hypothesis that the loss of H3K27me3 is linked to the inactivation of BRD4 in this case. YAP1 and Filamine-A IHC, commonly used in the setting of the molecular subgroups of medulloblastomas) [6], were performed and showed loss of expression in tumor cells (Fig. 3), with unknown clinical significance in this tumor type. Finally, regarding morphological specificities, as observed in CNS tumor with BCOR ITD, giant, “bizarre” cells with marked cytonuclear pleomorphism associated with geographical necrosis could be helpful to distinguish this tumor type. In addition to these features, large area of intratumoral desmoplasia and calcifications could be specific to this subgroup. These rare cases of CNS embryonal tumor characterized by BRD4–LEUTX fusion need more clinical integration associated with prognosis to lead to an integration into the CNS WHO classification.
To conclude, according to the 2021 WHO classification, CNS embryonal tumors are “a work in progress” due to the emergence of a classification based on molecular characteristics [12]. Our case report suggested a new subgroup of CNS embryonal tumor with high aggressive potential, BRD4–LEUTX fusion, loss of H3K27me3 protein expression, named “Embryonal CNS tumor with BRD4–LEUTX fusion”. This subgroup should be considered into the new CNS WHO classification if more cases reports allow us to better characterized this potential subgroup.
Abbreviations
- ITD
Internal tandem duplication
- CT
Computed tomography
- ATRT
Atypical teratoid rhabdoid tumor
- CNS
Central nervous system
- CNV
Copy number variations
- MRI
Magnetic resonance imaging
- NGS
Next generation sequencing
- WHO
World Health Organization
- IHC
Immunohistochemical
- HE
Hematoxylin–eosin
- DW
Diffusion-weighted
- ADC
Apparent diffusion coefficient
- ASL
Arterial spin labeling
- CBF
Cerebral blood flow
- BET
Bromodomain and extraterminal
Author contributions
LL, SAD, VL, MT, CR, CF, OD, NG, PD, PC compiled the radiological, surgical and clinical data; LL, IS, SAD conducted the neuropathological examinations; FE, CVC, CAM conducted the molecular studies; LL, SAD, IS, VL, MR, CF, CR, PC drafted the manuscript; all the authors reviewed the manuscript.
Funding
This work was performed with the support of grants awarded by the “Fonds Erasme” for Medical Research (Brussels, Belgium) via the Félicien Tomme Convention and by funding from the “Fonds Yvonne Boël” (Brussels, Belgium).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Declarations
Ethics approval and consent to participate
According to Belgian law, no written informed consent was required for archival material. The ethical committee thus waived the requirement for written informed consent from the participants.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Alexandrescu S, Meredith DM, Lidov HG, Alaggio R, Novello M, Ligon KL, Vargas SO. Loss of histone H3 trimethylation on lysine 27 and nuclear expression of transducin-like enhancer 1 in primary intracranial sarcoma, DICER1-mutant. Histopathology. 2021;78:265–275. doi: 10.1111/his.14217. [DOI] [PubMed] [Google Scholar]
- 2.Barresi S, Giovannoni I, Rossi S, Stracuzzi A, Quacquarini D, Cafferata B, Piscitelli D, De Leonardis F, Marzullo A, Alaggio R. A novel BRD4–LEUTX fusion in a pediatric sarcoma with epithelioid morphology and diffuse S100 expression. Genes Chromosomes Cancer. 2021;60:647–652. doi: 10.1002/gcc.22974. [DOI] [PubMed] [Google Scholar]
- 3.Capper D, Jones DTW, Sill M, Hovestadt V, Schrimpf D, Sturm D, Koelsche C, Sahm F, Chavez L, Reuss DE, Kratz A, Wefers AK, Huang K, Pajtler KW, Schweizer L, Stichel D, Olar A, Engel NW, Lindenberg K, Harter PN, Braczynski AK, Plate KH, Dohmen H, Garvalov BK, Coras R, Hölsken A, Hewer E, Bewerunge-Hudler M, Schick M, Fischer R, Beschorner R, Schittenhelm J, Staszewski O, Wani K, Varlet P, Pages M, Temming P, Lohmann D, Selt F, Witt H, Milde T, Witt O, Aronica E, Giangaspero F, Rushing E, Scheurlen W, Geisenberger C, Rodriguez FJ, Becker A, Preusser M, Haberler C, Bjerkvig R, Cryan J, Farrell M, Deckert M, Hench J, Frank S, Serrano J, Kannan K, Tsirigos A, Brück W, Hofer S, Brehmer S, Seiz-Rosenhagen M, Hänggi D, Hans V, Rozsnoki S, Hansford JR, Kohlhof P, Kristensen BW, Lechner M, Lopes B, Mawrin C, Ketter R, Kulozik A, Khatib Z, Heppner F, Koch A, Jouvet A, Keohane C, Mühleisen H, Mueller W, Pohl U, Prinz M, Benner A, Zapatka M, Gottardo NG, Driever PH, Kramm CM, Müller HL, Rutkowski S, von Hoff K, Frühwald MC, Gnekow A, Fleischhack G, Tippelt S, Calaminus G, Monoranu C-M, Perry A, Jones C, Jacques TS, Radlwimmer B, Gessi M, Pietsch T, Schramm J, Schackert G, Westphal M, Reifenberger G, Wesseling P, Weller M, Collins VP, Blümcke I, Bendszus M, Debus J, Huang A, Jabado N, Northcott PA, Paulus W, Gajjar A, Robinson GW, Taylor MD, Jaunmuktane Z, Ryzhova M, Platten M, Unterberg A, Wick W, Karajannis MA, Mittelbronn M, Acker T, Hartmann C, Aldape K, Schüller U, Buslei R, Lichter P, Kool M, Herold-Mende C, Ellison DW, Hasselblatt M, Snuderl M, Brandner S, Korshunov A, von Deimling A, Pfister SM. DNA methylation-based classification of central nervous system tumours. Nature. 2018;555:469–474. doi: 10.1038/nature26000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Decock A, Creytens D, Lefever S, Van der Meulen J, Anckaert J, De Ganck A, Deleu J, De Wilde B, Fierro C, Kuersten S, Luypaert M, Rottiers I, Schroth GP, Steyaert S, Vanderheyden K, VandenEynde E, Verniers K, Verreth J, Van Dorpe J, Vandesompele J. mRNA capture sequencing and RT-qPCR for the detection of pathognomonic, novel, and secondary fusion transcripts in FFPE tissue: a sarcoma showcase. Int J Mol Sci. 2022;23:11007. doi: 10.3390/ijms231911007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.D’Haene N, Meléndez B, Blanchard O, De Nève N, Lebrun L, Van Campenhout C, Salmon I. Design and validation of a gene-targeted, next-generation sequencing panel for routine diagnosis in gliomas. Cancers. 2019 doi: 10.3390/cancers11060773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ellison DW, Dalton J, Kocak M, Nicholson SL, Fraga C, Neale G, Kenney AM, Brat DJ, Perry A, Yong WH, Taylor RE, Bailey S, Clifford SC, Gilbertson RJ. Medulloblastoma: clinicopathological correlates of SHH, WNT, and non-SHH/WNT molecular subgroups. Acta Neuropathol. 2011;121:381–396. doi: 10.1007/s00401-011-0800-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henssen A, Thor T, Odersky A, Heukamp L, El-Hindy N, Beckers A, Speleman F, Althoff K, Schäfers S, Schramm A, Sure U, Fleischhack G, Eggert A, Schulte JH. BET bromodomain protein inhibition is a therapeutic option for medulloblastoma. Oncotarget. 2013;4:2080–2095. doi: 10.18632/oncotarget.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu W, Wang J, Yuan L, Zhang X, Ji Y, Song C, et al. Case report: a unique case of pediatric central nervous system embryonal tumor harboring the CIC–LEUTX fusion, germline NBN variant and somatic TSC2 mutation: expanding the spectrum of CIC-rearranged neoplasia. Front Oncol. 2 déc. 10.3389/fonc.2020.598970 [DOI] [PMC free article] [PubMed]
- 9.Ishi Y, Zhang Y, Zhang A, Sasaki T, Piunti A, Suri A, Watanabe J, Abe K, He X, Katagi H, Bhalla P, Natsumeda M, Zou L, Shilatifard A, Hashizume R. Therapeutic targeting of EZH2 and BET BRD4 in pediatric rhabdoid tumors. Mol Cancer Ther. 2022;21:715–726. doi: 10.1158/1535-7163.MCT-21-0646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jouhilahti E-M, Madissoon E, Vesterlund L, Töhönen V, Krjutškov K, Plaza Reyes A, Petropoulos S, Månsson R, Linnarsson S, Bürglin T, Lanner F, Hovatta O, Katayama S, Kere J. The human PRD-like homeobox gene LEUTX has a central role in embryo genome activation. Development. 2016;143:3459–3469. doi: 10.1242/dev.134510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lebrun L, Meléndez B, Blanchard O, De Nève N, Van Campenhout C, Lelotte J, Balériaux D, Riva M, Brotchi J, Bruneau M, De Witte O, Decaestecker C, D’Haene N, Salmon I. Clinical, radiological and molecular characterization of intramedullary astrocytomas. Acta Neuropathol Commun. 2020;8:128. doi: 10.1186/s40478-020-00962-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Louis DN, Perry A, Wesseling P, Brat DJ, Cree IA, Figarella-Branger D, Hawkins C, Ng HK, Pfister SM, Reifenberger G, Soffietti R, von Deimling A, Ellison DW. The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro Oncol. 2021;23:1231–1251. doi: 10.1093/neuonc/noab106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maury E, Hashizume R. Epigenetic modification in chromatin machinery and its deregulation in pediatric brain tumors: insight into epigenetic therapies. Epigenetics. 2017;12:353–369. doi: 10.1080/15592294.2016.1278095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meredith DM, Alexandrescu S. Embryonal and non-meningothelial mesenchymal tumors of the central nervous system—advances in diagnosis and prognostication. Brain Pathol. 2022;32:e13059. doi: 10.1111/bpa.13059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moreno V, Saluja K, Pina-Oviedo S. NUT carcinoma: clinicopathologic features, molecular genetics and epigenetics. Front Oncol. 2022;12:860830. doi: 10.3389/fonc.2022.860830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roosen M, Odé Z, Bunt J, Kool M. The oncogenic fusion landscape in pediatric CNS neoplasms. Acta Neuropathol. 2022;143:427–451. doi: 10.1007/s00401-022-02405-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sturm D, Orr BA, Toprak UH, Hovestadt V, Jones DTW, Capper D, Sill M, Buchhalter I, Northcott PA, Leis I, Ryzhova M, Koelsche C, Pfaff E, Allen SJ, Balasubramanian G, Worst BC, Pajtler KW, Brabetz S, Johann PD, Sahm F, Reimand J, Mackay A, Carvalho DM, Remke M, Phillips JJ, Perry A, Cowdrey C, Drissi R, Fouladi M, Giangaspero F, Łastowska M, Grajkowska W, Scheurlen W, Pietsch T, Hagel C, Gojo J, Lötsch D, Berger W, Slavc I, Haberler C, Jouvet A, Holm S, Hofer S, Prinz M, Keohane C, Fried I, Mawrin C, Scheie D, Mobley BC, Schniederjan MJ, Santi M, Buccoliero AM, Dahiya S, Kramm CM, von Bueren AO, von Hoff K, Rutkowski S, Herold-Mende C, Frühwald MC, Milde T, Hasselblatt M, Wesseling P, Rößler J, Schüller U, Ebinger M, Schittenhelm J, Frank S, Grobholz R, Vajtai I, Hans V, Schneppenheim R, Zitterbart K, Collins VP, Aronica E, Varlet P, Puget S, Dufour C, Grill J, Figarella-Branger D, Wolter M, Schuhmann MU, Shalaby T, Grotzer M, van Meter T, Monoranu C-M, Felsberg J, Reifenberger G, Snuderl M, Forrester LA, Koster J, Versteeg R, Volckmann R, van Sluis P, Wolf S, Mikkelsen T, Gajjar A, Aldape K, Moore AS, Taylor MD, Jones C, Jabado N, Karajannis MA, Eils R, Schlesner M, Lichter P, von Deimling A, Pfister SM, Ellison DW, Korshunov A, Kool M. New brain tumor entities emerge from molecular classification of CNS-PNETs. Cell. 2016;164:1060–1072. doi: 10.1016/j.cell.2016.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taylor MD, Northcott PA, Korshunov A, Remke M, Cho Y-J, Clifford SC, Eberhart CG, Parsons DW, Rutkowski S, Gajjar A, Ellison DW, Lichter P, Gilbertson RJ, Pomeroy SL, Kool M, Pfister SM. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol. 2012;123:465–472. doi: 10.1007/s00401-011-0922-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wong M, Mayoh C, Lau LMS, Khuong-Quang D-A, Pinese M, Kumar A, Barahona P, Wilkie EE, Sullivan P, Bowen-James R, Syed M, Martincorena I, Abascal F, Sherstyuk A, Bolanos NA, Baber J, Priestley P, Dolman MEM, Fleuren EDG, Gauthier M-E, Mould EVA, Gayevskiy V, Gifford AJ, Grebert-Wade D, Strong PA, Manouvrier E, Warby M, Thomas DM, Kirk J, Tucker K, O’Brien T, Alvaro F, McCowage GB, Dalla-Pozza L, Gottardo NG, Tapp H, Wood P, Khaw S-L, Hansford JR, Moore AS, Norris MD, Trahair TN, Lock RB, Tyrrell V, Haber M, Marshall GM, Ziegler DS, Ekert PG, Cowley MJ. Whole genome, transcriptome and methylome profiling enhances actionable target discovery in high-risk pediatric cancer. Nat Med. 2020;26:1742–1753. doi: 10.1038/s41591-020-1072-4. [DOI] [PubMed] [Google Scholar]
- 20.Wu S-Y, Lee A-Y, Lai H-T, Zhang H, Chiang C-M. Phospho switch triggers BRD4 chromatin binding and activator recruitment for gene-specific targeting. Mol Cell. 2013;49:843–857. doi: 10.1016/j.molcel.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu T, Kamikawa YF, Donohoe ME. Brd4’s bromodomains mediate histone H3 acetylation and chromatin remodeling in pluripotent cells through P300 and Brg1. Cell Rep. 2018;25:1756–1771. doi: 10.1016/j.celrep.2018.10.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.