Abstract
Hyperlactatemia often occurs in critically ill patients during severe sepsis/septic shock and is a powerful predictor of mortality. Lactate is the end product of glycolysis. While hypoxia due to inadequate oxygen delivery may result in anaerobic glycolysis, sepsis also enhances glycolysis under hyperdynamic circulation with adequate oxygen delivery. However, the molecular mechanisms involved are not fully understood. Mitogen-activated protein kinase (MAPK) families regulate many aspects of the immune response during microbial infections. MAPK phosphatase (MKP)-1 serves as a feedback control mechanism for p38 and JNK MAPK activities via dephosphorylation. Here, we found that mice deficient in Mkp-1 exhibited substantially enhanced expression and phosphorylation of 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase (PFKFB) 3, a key enzyme that regulates glycolysis following systemic Escherichia coli infection. Enhanced PFKFB3 expression was observed in a variety of tissues and cell types, including hepatocytes, macrophages, and epithelial cells. In bone marrow–derived macrophages, Pfkfb3 was robustly induced by both E. coli and lipopolysaccharide, and Mkp-1 deficiency enhanced PFKFB3 expression with no effect on Pfkfb3 mRNA stability. PFKFB3 induction was correlated with lactate production in both WT and Mkp-1−/− bone marrow–derived macrophage following lipopolysaccharide stimulation. Furthermore, we determined that a PFKFB3 inhibitor markedly attenuated lactate production, highlighting the critical role of PFKFB3 in the glycolysis program. Finally, pharmacological inhibition of p38 MAPK, but not JNK, substantially attenuated PFKFB3 expression and lactate production. Taken together, our studies suggest a critical role of p38 MAPK and MKP-1 in the regulation of glycolysis during sepsis.
Keywords: infection, p38 MAPK, c-Jun N-terminal kinase, dual-specificity phosphoprotein phosphatase, glycolysis, sepsis
Abbreviations: BMDM, bone marrow–derived macrophage; F-2,6-bisP, fructose-2,6-bisphosphate; HK, hexokinase; IHC, immunohistochemistry; LPS, lipopolysaccharide; MAPK, mitogen-activated protein kinase; MAPKAPK, MAPK-activated protein kinase; MKP, MAPK phosphatase; PFK, phosphofructokinase; PFKFB, phosphofructo-2-kinase/fructose-2,6-biphosphatase; qRT-PCR, quantitative RT-PCR
Lactic acidosis is common in critically ill patients with severe sepsis/septic shock. In fact, a serum lactate level greater than 2 mM is one of the criteria for diagnosis of septic shock by the Sepsis Definition Task Force (1, 2). Serum lactate is also a powerful predictor of mortality in patients with sepsis (3, 4), and a serum lactate level greater than 4 mM is associated with a mortality rate of 28.4% (3). Although hyperlactatemia has been considered an indication of anaerobic glycolysis within tissues due to oxygen “debt” at a cellular level, the theory of inadequate tissue oxygenation does not explain many clinical or experimental observations. Septic patients usually exhibit increased oxygen transport, particularly after hemodynamic resuscitation and oxygen delivery (5, 6, 7), and yet hyperlactatemia persists in a subset of sepsis patients. Additionally, lactic acidosis can develop without tissue hypoxia in a variety of tissues, including muscle, intestinal mucosa, heart, lung, and brain (8, 9, 10, 11). More recent studies support the idea that sepsis promotes a marked change in the metabolic program, shifting the glucose metabolic pathway from oxidative phosphorylation to glycolysis (12, 13, 14, 15). The metabolic reprogramming of glycolysis is an important mechanism that regulates immune cell activation mediated by innate immune sensors/receptors (16, 17).
Glycolysis consists of 10 consecutive enzymatic reactions (18). The three key, rate-limiting enzymes of glycolysis are hexokinase (HK), phosphofructokinase (PFK) 1, and pyruvate kinase. HK converts glucose to glucose-6-phosphate and PFK1 catalyzes the important “committed” step of glycolysis, the conversion of fructose-6-phosphate to fructose-1,6-bisphosphate. Fructose-2,6-bisphosphate (F-2,6-bisP), the most potent allosteric activator of PFK1, is produced from fructose-6-phosphate by a family of bifunctional 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase (PFKFB) enzymes and broken down by the same class of enzymes. Among the PFKFB family members, the inducible PFKFB3 has the highest kinase-activity/phosphatase-activity ratio, thus it is able to produce high levels of F-2,6-bisP. Additionally, PFKFB3 can be further activated by a number of protein kinases via phosphorylation (19, 20, 21, 22). PFKFB3 activation dramatically bolsters PFK1 activity and PFK1-mediated glycolysis. While the role of PFKFB3 in glycolysis is well studied (23, 24), the regulation of PFKFB3 during the immune response is still not fully understood. It has been shown that HIF1α and NF-κB, two major transcription factors, can regulate Pfkfb3 gene expression and glycolysis (25, 26). In the present study, we found that the absence of MKP-1, a crucial negative regulator of p38 and JNK MAPKs, substantially elevated PFKFB3 expression and phosphorylation both in vivo, in Escherichia coli–infected septic mice, and in vitro, in lipopolysaccharide (LPS)-stimulated macrophages. The induction and phosphorylation of PFKFB3 by LPS in macrophages were primarily mediated by p38 MAPK. Pharmacological inhibition of PFKFB3 or p38 MAPK markedly attenuated lactate production by macrophages following inflammatory stimuli. These results indicate that p38 MAPK is a crucial initiator of glycolysis during sepsis, and MKP-1 serves as a feedback control mechanism of sepsis-induced glycolysis. Our studies suggest that p38 MAPK and MKP-1 are important regulators of glycolytic reprogramming during sepsis.
Results
Mkp-1−/− mice exhibit significantly elevated PFKFB3 expression relative to WT mice when infected with E. coli
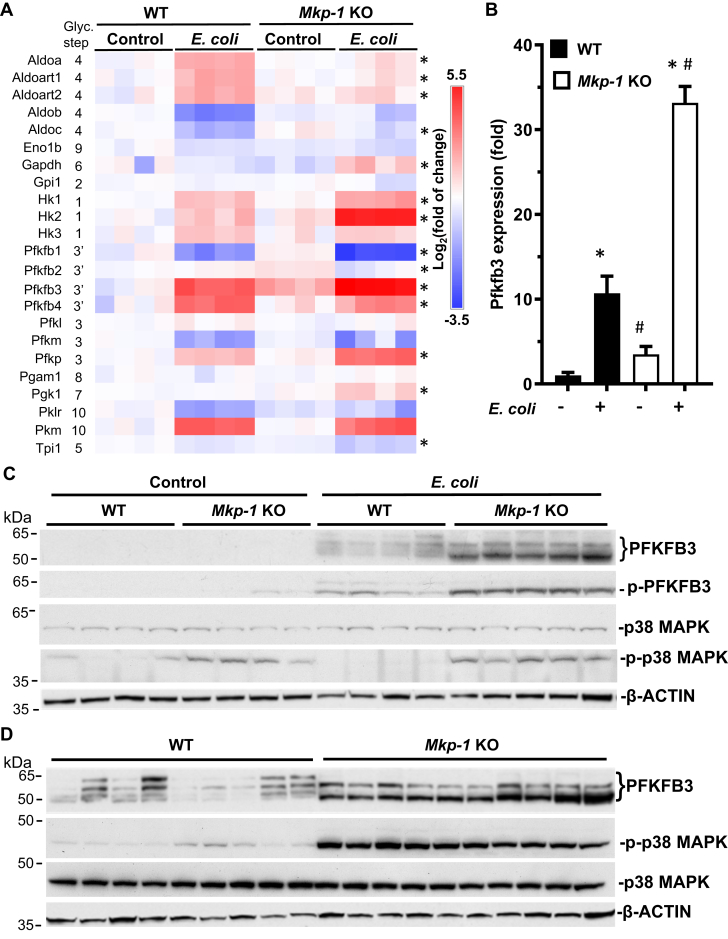
To understand the molecular basis of increased glycolysis following sepsis, we examined the RNA-seq database generated from liver samples of PBS-treated and E. coli–infected WT and Mkp-1 KO mice (27). We analyzed the gene expression levels of the enzymes that catalyze the 10 steps of glycolysis and found that E. coli infection increased the mRNA levels of several glycolytic enzymes, including HK1-3, PFKFB3, PFKFB4, PFKP, and PKM (Fig. 1A). Interestingly, Mkp-1 deficiency substantially exacerbated the expression of three key glycolytic enzymes: HK2, PFKFB3, and PFKP. Because PFKP is a platelet-specific enzyme (28), its expression likely reflects the platelet contents in the liver tissues. Although Hk2 mRNA levels were substantially higher in the livers of E. coli–infected Mkp-1 KO mice, HK2 protein levels did not significantly differ from those of WT mice (data not shown). PFKFB3 is particularly interesting since PFKFB3 generates F-2,6-BisP, an allosteric activator of the rate-limiting enzyme PFK1. Thus, enhanced PFKFB3 activity is expected to stimulate glycolysis substantially. Mkp-1 deficiency significantly enhanced Pfkfb3 mRNA expression levels in the livers of Mkp-1 KO mice both in controls and after E. coli infection (Fig. 1B). We then examined the levels of liver PFKFB3 protein in the control and E. coli–infected mice by Western blotting using a polyclonal PFKFB3 antibody (Fig. 1C). PFKFB3 protein levels were very low in the livers of both control WT and Mkp-1 KO mice. E. coli infection increased the amount of 3 to 4 PFKFB3 isoforms of 52 to 67 kDa in WT mice, and PFKFB3 protein levels were significantly higher in E. coli–infected Mkp-1 KO mice than in E. coli–infected WT mice (Fig. 1, C and D). Moreover, the relative abundances of the PFKFB3 isoforms expressed in E. coli–infected Mkp-1 KO mice were also different from those in E. coli–infected WT mice. Unlike in the WT mice, where the most abundant PFKFB3 isoforms were 61 and 67 kDa, respectively, E. coli–infected Mkp-1 KO mice preferentially expressed the 52 and 61 kDa PFKFB3 isoforms. The 52 kDa PFKFB3 isoform was substantially increased in E. coli–infected Mkp-1 KO mice relative to E. coli–infected WT mice.
Figure 1.
Mkp-1 deficiency enhances Pfkfb3 expression in Escherichia coli–infected mice. WT and Mkp-1 KO mice on a C57/129 background were infected with E. coli (O55:B5) i.v. at a dose of 2.5 × 107 CFU/g b.w. or injected with PBS (controls). Mice were euthanized after 24 h and total RNA was isolated from the livers using Trizol for RNA-seq analyses. The livers were homogenized to extract soluble protein for Western blot analysis. A, heatmap of the mRNA levels of glycolytic enzymes. The copy numbers of RNA transcripts for each gene were normalized to the average number in WT controls to calculate fold change. When the transcript number is 0 for a given gene in a specific animal, we gave an arbitrary number that is lower than the lowest value in that group. These values were log2-transformed to generate the heatmap. Each column represents a distinct animal. ∗p < 0.05, comparing E. coli–infected Mkp-1 KO and E. coli–infected WT mice (t test, n = 4). B, Pfkfb3 mRNA levels in the livers of PBS-treated or E. coli–infected mice determined by RNA-seq. Data are shown as means ± S.E (n = 4 mice in each group). ∗p < 0.05, compared to PBS-treated mice of the same genotype; #, p < 0.05, compared to WT mice of the same treatment (t test, n = 4). C, levels of total and phosphorylated PFKFB3 as well as total and phosphorylated p38 MAPK in the livers of control and E. coli–infected mice. Total and phosphorylated PFKFB3 and p38 MAPK were detected by Western blotting using an antibody against total PFKFB3, Ser461-phosphorylated PFKFB3 (p-PFKFB3), total p38 MAPK, and Thr180/Tyr182-phosphorylated p38 MAPK. Comparable protein loading was verified by Western blotting with β-ACTIN antibody (Lower panel). D, side-by-side comparison of total PFKFB3, Thr180/Tyr182-phosphorylated p38 MAPK, and total p38 MAPK levels in liver samples of large groups of E. coli–infected WT and Mkp-1 KO mice. Each lane was from a distinct animal. Representative Western blotting results are shown. MAPK, mitogen-activated protein kinase; MKP, MAPK phosphatase; PFKFB, phosphofructo-2-kinase/fructose-2,6-biphosphatase.
Previous studies have shown that MAPK-activated protein kinase (MAPKAPK) 2, a downstream target of p38 MAPK (29, 30), phosphorylates PFKFB3 at Ser461, resulting in elevated enzymatic activity (22). Since MKP-1 negatively regulates p38 MAPK and MAPKAPK-2 activity (31), we assessed Ser461 phosphorylation in PFKFB3 protein by Western blot analysis using a phospho-specific PFKFB3 antibody (Fig. 1C). Similar to the total PFKFB3 protein abundance, Ser461-phosphorylated PFKFB3 levels were substantially higher in E. coli–infected Mkp-1 KO mice than in E. coli–infected WT mice. We also assessed the level of p38 MAPK activity by Western blotting using antibodies against the Thr180/Tyr182-phosphorylated and total p38 MAPK. Compared to WT mice, the liver p38 MAPK activity levels, indicated by p-p38 MAPK levels, were markedly higher in Mkp-1 KO mice, particularly after E. coli infection (Fig. 1C). Comparison of PFKFB3 levels in larger groups of samples (n = 9–10) confirmed the elevated PFKFB3 expression in the livers of E. coli–infected Mkp-1 KO mice relative to similarly infected WT mice (Fig. 1D).
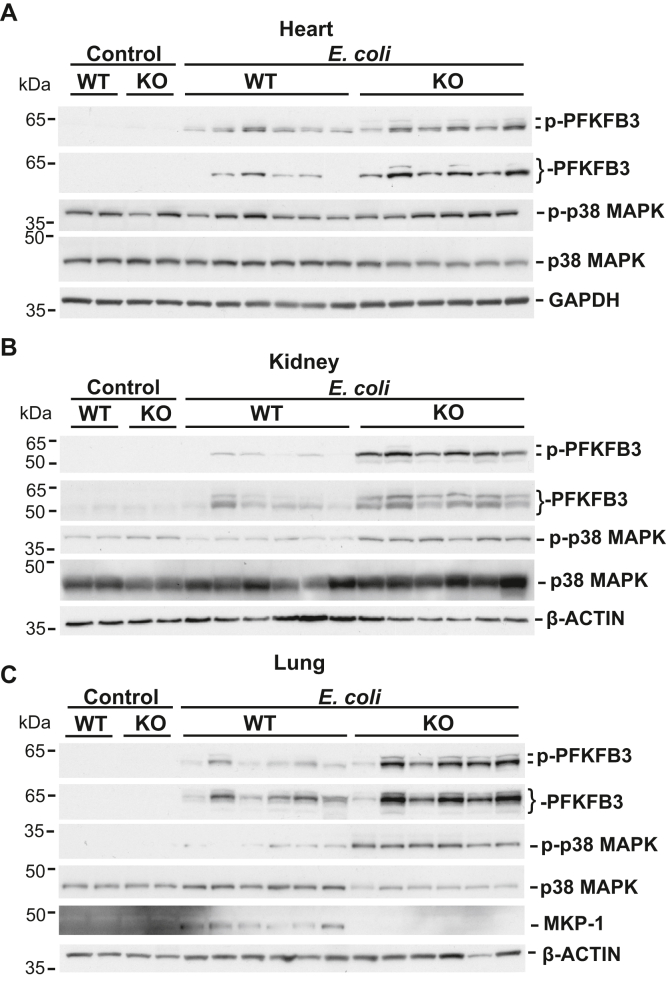
The myocardium utilizes free fatty acids as its primary energy source but switches to aerobic glycolysis during sepsis (32). Sepsis can induce an early anabolic response in renal tissue that is characterized by a shift of metabolism toward aerobic glycolysis (33). The lungs can create lactate through glycolysis during acute lung injury without tissue hypoxia (34). Lactate is metabolized primarily by the liver and, to some extent, by the kidneys (5). We analyzed the effect of Mkp-1 deficiency on PFKFB3 expression in these major organs both before and after E. coli infection. Greater abundance of total PFKFB3 and Ser461-phosphorylated PFKFB3 proteins were observed in the hearts, kidneys, and lungs of E. coli–infected Mkp-1 KO mice in comparison to the similarly infected WT mice (Fig. 2). p38 MAPK activity was also higher in the kidneys and lungs of E. coli–infected Mkp-1 KO mice than in those of similarly infected WT mice (Fig. 2, B and C). MKP-1 protein was detected in the lungs of E. coli–infected WT mice, but not in Mkp-1 KO mice (Fig. 2C). MKP-1 was not detected in the lungs of control WT mice, likely due to its low steady state level. It has been shown that the lung is the organ with the highest Mkp-1 expression (35). MKP-1 protein was not detected in the livers, hearts, and kidneys of WT mice either with or without E. coli infection, likely due to the low or transient MKP-1 expression in these organs.
Figure 2.
Mkp-1 deficiency exacerbates PFKFB3 induction in Escherichia coli–infected mice. WT and Mkp-1 KO mice were infected with E. coli (O55:B5) or injected with PBS (controls) as in Figure 1. Mice were euthanized after 24 h. Hearts, kidneys, and lungs were harvested to extract soluble protein for Western blot analysis. Total and phosphorylated PFKFB3, and total and phosphorylated p38 MAPK in the hearts (A), kidneys (B), and lungs (C) of PBS-treated (control) and E. coli–infected WT and Mkp-1 KO mice were detected by Western blotting. The lung samples were also analyzed by Western blotting to detect MKP-1 expression, using a rabbit mAb (C). A mAb against GAPDH or β-ACTIN was used to control for sample loading. Each lane was from a distinct animal. Representative Western blotting results are shown. MAPK, mitogen-activated protein kinase; MKP, MAPK phosphatase; PFKFB, phosphofructo-2-kinase/fructose-2,6-biphosphatase.
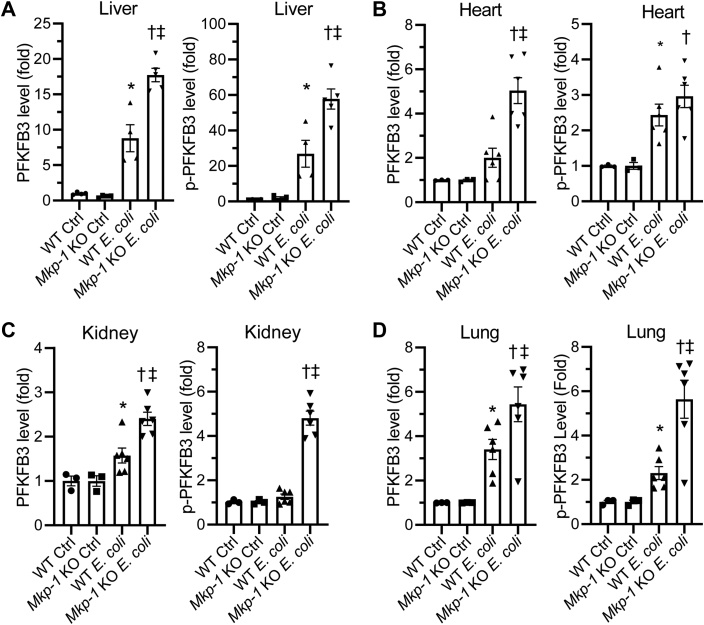
To quantify the levels of total and Ser461-phosphorylated PFKFB3, we performed densitometry analysis. Densitometry analysis confirmed significant differences in both total and Ser461-phosphorylated PFKFB3 protein in the livers, kidneys, and lungs between E. coli–infected WT and Mkp-1 KO mice (Fig. 3, A, C, and D). In the heart, total PFKFB3 protein levels were also significantly higher in E. coli–infected Mkp-1 KO mice than similarly infected WT mice (Fig. 3B). Although the levels of Ser461-phosphorylated PFKFB3 protein in the hearts appeared higher in E. coli–infected Mkp-1 KO mice than similarly infected WT mice, the difference was not statistically significant (Fig. 3B).
Figure 3.
Quantification of total and Ser461-phosphorylated PFKFB3 in different organs of PBS-treated (control) and Escherichia coli–infected WT and Mkp-1 KO mice. Total (Left graph) and Ser461-phosphorylated (Right graph) PFKFB3 in livers (A), hearts (B), kidneys (C), and lungs (D) were detected by Western blotting. The expression was quantified by densitometry and normalized to a house-keeping protein. The levels were expressed as fold relative to the mean of WT control. ∗p < 0.05, compared to WT control; †, p < 0.05, compared to Mkp-1 KO control; ‡, p < 0.05, compared to E. coli–infected WT mice (t test, n = 3–6). MKP, MAPK phosphatase; PFKFB, phosphofructo-2-kinase/fructose-2,6-biphosphatase.
PFKFB3 was more abundantly induced as a nuclear protein by E. coli infection in Mkp-1 KO mice in a variety of cell types
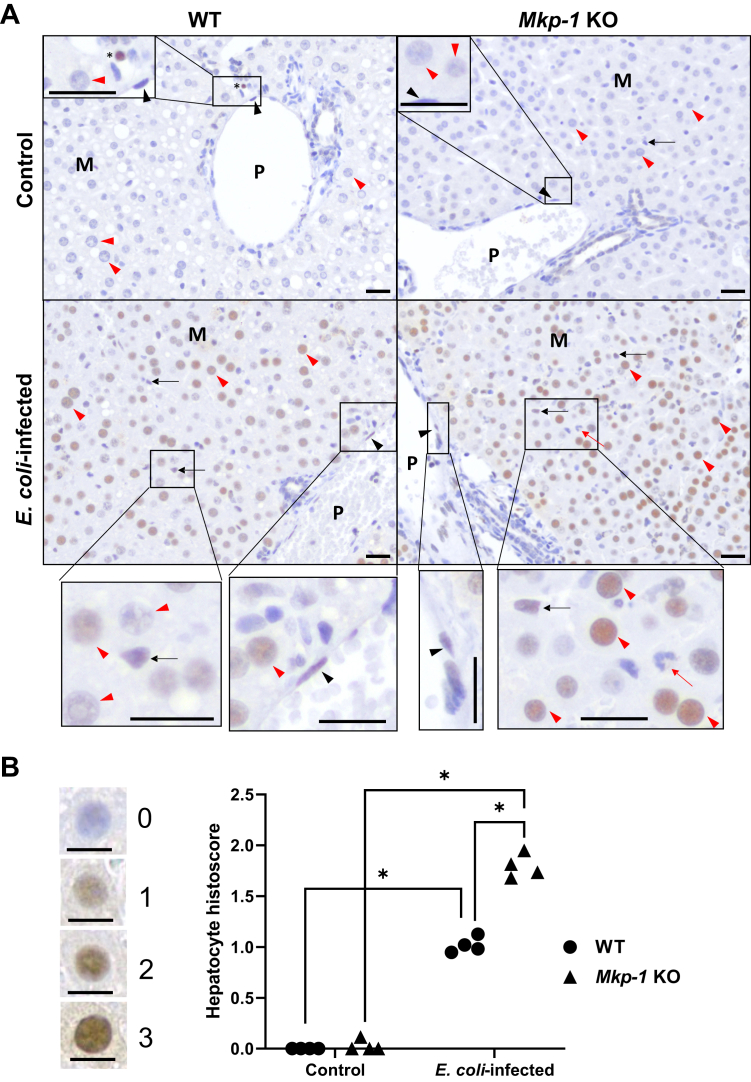
To understand the cell types and subcellular localization of PFKFB3, we performed immunohistochemistry (IHC) on tissues harvested from control and E. coli–infected WT and Mkp-1 KO mice, specifically the liver, spleen, kidney, heart, and lungs. Negative controls of the IHC experiment performed with tissues of E. coli–infected Mkp-1 KO mice without using the primary PFKFB3 antibody yielded no staining in the liver, heart, kidney, lung, adipose, and spleen tissues (Fig. S1), highlighting the specificity of the assays. In the livers of both control WT and Mkp-1 KO mice, PFKFB3 protein expression was very low (Fig. 4A, Upper panels). Hepatocyte PFKFB3 staining was largely negative, although PFKFB3 staining was occasionally seen in intravascular leukocytes (Fig. 4A, Upper left panel: control WT, marked by an asterisk). In WT mice, upon E. coli infection, PFKFB3 protein staining was moderately increased markedly in the nuclei of various cell types, most clearly seen in hepatocytes (marked by red arrowheads) in all regions of the hepatic lobules, including the portal region, midzone, and centrilobular region (not shown) (Fig. 4A, Lower left panel). Weakly positive PFKFB3 staining was seen occasionally in hepatic resident macrophages (often referred to as Kupffer cells, marked by black arrow) and large vessel endothelial cells (marked by black arrowhead) (Fig. 4A, Lower left panel). PFKFB3 staining was markedly stronger in the livers in E. coli–infected Mkp-1 KO mice than in E. coli–infected WT mice, particularly in the hepatocytes in all regions of the hepatic lobules. However, staining was less abundant in vascular endothelial cells and Kupffer cells (Fig. 4A, Lower right panel). Rarely, intravascular leukocytes, likely monocytes, also stained strongly (data not shown). In contrast, neutrophils (marked by a red arrow in E. coli–infected Mkp-1 KO mice, Figure 4A, Lower right panel) appeared to have very low PFKFB3 expression, essentially undetectable by IHC. These findings support the notion that the increases in PFKFB3 protein levels detected via immunoblotting are primarily due to increased hepatocyte PFKFB3 expression.
Figure 4.
PFKFB3 is strongly induced in response to Escherichia coli infection in hepatocytes. WT and Mkp-1 KO mice (C57BL6/J) were infected i.v. with E. coli at a dose of 1 × 107 CFU/g b.w. and euthanized 24 h postinfection. The livers were excised, fixed in formalin, and sectioned for IHC with a rabbit polyclonal antibody against mouse PFKFB3. After immunohistochemical staining, the sections were counterstained with hematoxylin. A, increased PFKFB3 expression in the liver of E. coli–infected Mkp-1 KO mice compared to WT mice. Note the marked expression in the nuclei of hepatocytes (red arrowhead), Kupffer cells (black arrow), and endothelial cells (black arrowhead) in E. coli–infected WT and Mkp-1 KO mice. P: portal vein; M: midzonal region; Red arrow: neutrophil; ∗: intravascular leukocytes. Black bar length in all images: 20 μm. B, histoscores of PFKFB3 staining in the hepatocytes of control and E. coli–infected WT and Mkp-1 KO mice. Four representative immunohistochemical fields (each covering 0.03 mm2) were obtained using Aperio ImageScope software for histoscore analysis. Immunohistochemical staining of every hepatocyte nucleus was graded as follows: 0, nonstaining; 1, weak; 2, moderate; or 3, strong (shown on the left side of the graph). The mean histoscore of each field was calculated from 50 to 100 hepatocytes. Each dot in the graph represents the mean hepatocyte histoscore from a representative field. The histoscores were compared by two-way ANOVA. ∗p < 0.05 (n = 4). Black bar length in all images: 10 μm. MKP, MAPK phosphatase; PFKFB, phosphofructo-2-kinase/fructose-2,6-biphosphatase; IHC, immunohistochemistry.
To further quantify the PFKFB3 staining differences in the hepatocytes of WT and Mkp-1 KO mice with or without E. coli infection, we performed a semiquantitative histoscore analysis based on the intensity of hepatocyte nuclear PFKFB3 staining. Representative hepatocyte nuclei and associated scores are shown on the left side of Figure 4B. Histoscore analysis indicates that hepatocyte PFKFB3 expression was markedly increased after E. coli infection in both WT and Mkp-1 KO mice (Fig. 4B). E. coli–induced hepatocyte PFKFB3 expression was significantly higher in Mkp-1 KO mice than in WT mice (Fig. 4B).
In the spleens of control WT and Mkp-1 KO mice, strong PFKFB3 staining was present in the nuclei of isolated leukocytes (marked by black arrowheads), presumably lymphocytes or dendritic cells, in the white pulp (Fig. S2, Upper panels). Weak nuclear PFKFB3 staining was present in red pulp macrophages (marked by thin black arrows), identifiable by more abundant cytoplasm, reniform nuclei, and more open chromatin than lymphocytes. Upon E. coli infection, mild to moderate lymphocytolysis (lymphocyte apoptosis with nuclear condensation and fragmentation) was present in the white pulp of both WT and Mkp-1 KO mice (Fig. S2, Lower panels, marked in ovals). In E. coli–infected WT mice, nuclear-stained cells were seen rarely in the white pulp, similar to the level of uninfected controls. Occasionally, moderate PFKFB3 staining was seen in the nuclei of macrophages within the red pulp (Fig. S2, Lower left panel, thin black arrow). A larger number of marginal zone leukocytes and red pulp macrophages were stained strongly positive in the nuclei in E. coli–infected Mkp-1 KO mice than in E. coli–infected WT mice (Fig. S2, Lower right panel). Additionally, some lymphocytes in the red pulp of E. coli–infected Mkp-1 KO mice were also stained strongly positive in the nuclei, although their numbers were fewer than the positively stained red pulp macrophages. In infected mice, megakaryocytes, denoted by the abundant cytoplasm and multiple nuclei, had weak nuclear and cytoplasmic staining (Fig. S2, Lower right panel, red arrowhead). In all sections, the faint cytoplasmic brown pigment in resident macrophages (Fig. S2, thin red arrows) was probably a result of iron pigment due to hemosiderin accumulation, as confirmed with H & E staining (data not shown).
In the kidneys of uninfected WT mice, weak to moderate nuclear PFKFB3 staining was present in the proximal tubules, distal convoluted tubules, and collecting ducts (Fig. S3). In the kidneys of uninfected Mkp-1 KO mice, variable mild to moderate nuclear PFKFB3 staining was seen in the distal convoluted tubules and collecting ducts, and moderate nuclear PFKFB3 staining was present in some proximal tubules (Fig. S3). Upon E. coli infection, strong PFKFB3 staining was seen in the nuclei of collecting ducts in the cortex and medulla of WT mice (Fig. S3). Occasionally, moderate nuclear PFKFB3 staining was also seen in glomerular endothelial cells. Compared to E. coli–infected WT mice, the kidneys of E. coli–infected Mkp-1 KO mice had markedly stronger nuclear PFKFB3 staining, particularly in the proximal tubules, distal tubules, and collecting ducts (Fig. S3). Additionally, strong nuclear PFKFB3 staining was often seen in glomerular endothelial cells (marked by red arrow in glomerulus) (Fig. S3, inlets of E. coli-infected mice), vascular endothelial cells (not shown), and occasionally intravascular leukocytes (not shown).
In the heart, without E. coli infection, almost all cells were stained negative for PFKFB3 expression (Fig. S4, Upper panels). Upon E. coli infection, small to medium sized vascular endothelial cells (marked by thin arrow) were stained moderately positive in their nuclei, while interstitial cells, presumably capillary endothelial cells (marked by black arrowhead), were stained weakly to moderately positive in their nuclei in the WT mice (Fig. S4, Lower panels). Compared to the WT mice, nuclear PFKFB3 staining in vascular endothelial cells (thin arrow) was stronger in Mkp-1 KO mice (Fig. S4, Lower panels). Interstitial cells, likely capillary endothelial cells given their flattened nuclei (marked by black arrowhead, Fig. S4, Lower panels), were stained moderately to strongly positive in the hearts of Mkp-1 KO mice. Additionally, moderate PFKFB3 staining was seen in the nuclei of some circulating leukocytes (marked by red arrowhead) in the heart of E. coli–infected Mkp-1 KO mice (Fig. S4, Lower right panel). Interestingly, there was no detected expression of PFKFB3 within the cardiomyocytes in any of the mice.
Without infection, PFKFB3 staining was largely negative in the lungs of both WT and Mkp-1 KO mice (Fig. S5). After E. coli infection, nuclear PFKFB3 staining was occasionally seen in endothelial cells, and moderate PFKFB3 staining was occasionally seen in alveolar macrophages in WT mice. In Mkp-1 KO mice, strongly stained PFKFB3-positive cells, specifically alveolar macrophages and alveolar endothelial cells, were more prevalent in the lungs than in E. coli–infected WT mice (Fig. S5). Additionally, vascular endothelial cells and many pneumocytes were also stained moderately to strongly positive in E. coli–infected Mkp-1 KO mice (Fig. S5). Occasionally, circulating leukocytes had moderate nuclear labeling with PFKFB3 (marked by thin red arrow). Given the widespread PFKFB3 expression in macrophages detected via IHC, we decided to further characterize this relationship utilizing in vitro assays.
Mkp-1 deficiency enhances E. coli– or LPS-induced Pfkfb3 mRNA expression in macrophages without affecting Pfkfb3 mRNA stability
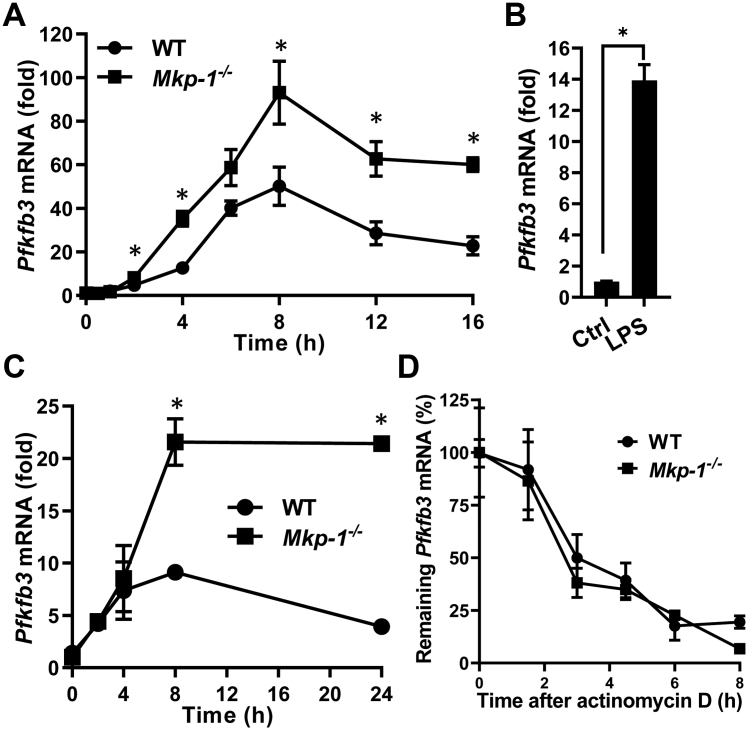
It has been shown that macrophages enhance glycolysis upon TLR ligand stimulation (16). To assess the effect of Mkp-1 deficiency on Pfkfb3 expression in macrophages, we examined the effects of heat-killed E. coli or LPS on Pfkfb3 expression in WT and Mkp-1−/− bone marrow–derived macrophage (BMDM) using quantitative RT-PCR (qRT-PCR). For two reasons, LPS was utilized to study the regulation of Pfkfb3 by MKP-1 during the cellular response to E. coli infection: 1) LPS, an endotoxin, is a critical part of the Gram-negative bacterium E. coli; and 2) LPS is a soluble TLR4 ligand. Thus, it stimulates cells uniformly and is easier to use than E. coli particles. Stimulation of macrophages with heat-killed E. coli elicited a dramatic increase in Pfkfb3 mRNA in a time-dependent manner (Fig. 5A). Pfkfb3 mRNA reached peak levels at approximately 8 h and then maintained high levels for up to 16 h. Similarly, LPS substantially induced the expression of Pfkfb3 mRNA in both WT and Mkp-1−/− BMDM (Fig. 5, B and C). Compared to WT BMDM, Mkp-1−/− BMDM expressed significantly greater levels of Pfkfb3 mRNA after both E. coli (Fig. 5A) and LPS stimulation (Fig. 5C).
Figure 5.
Mkp-1 deficiency enhances Pfkfb3 mRNA expression in macrophages following inflammatory stimuli but has little effect on Pfkfb3 mRNA stability.A, kinetics of Pfkfb3 mRNA induction following Escherichia coli stimulation. WT and Mkp-1−/− BMDM were stimulated with heat-killed E. coli (MOI: 10:1) for different times. Pfkfb3 mRNA levels in the samples were quantitated by qRT-PCR. The results were normalized to 18S ribosomal RNA. The expression of mRNA is presented as fold change relative to control cells. ∗p < 0.05, compared to WT at the same time-point (t test, n = 6). B, induction of Pfkfb3 mRNA in WT BMDM by LPS. WT BMDM were treated with LPS for 6 h. ∗p < 0.05, compared to WT control (t test, n = 6). C, kinetics of Pfkfb3 mRNA induction in WT and Mkp-1−/− BMDM following LPS stimulation. WT and Mkp-1−/− BMDM were stimulated with 100 ng/ml LPS for different times. ∗p < 0.05, compared to WT at the same time-point (t test, n = 3). D, the decay of Pfkfb3 mRNA in WT and Mkp-1−/− BMDM after actinomycin D treatment. WT and Mkp-1−/− BMDM were stimulated with 100 ng/ml LPS for 8 h and then treated with 10 ng/ml actinomycin D. Cells were harvested at different times to quantitate Pfkfb3 mRNA levels using qRT-PCR. The values were normalized to 18S ribosomal RNA and expressed as percentage relative to levels prior to actinomycin D addition. The half-life of Pfkfb3 mRNA in WT and Mkp-1−/− BMDM was estimated to be 2.84 and 2.33 h, respectively. MKP, MAPK phosphatase; PFKFB, phosphofructo-2-kinase/fructose-2,6-biphosphatase; BMDM, bone marrow–derived macrophage; LPS, lipopolysaccharide; qRT-PCR, quantitative RT-PCR.
Increased mRNA stability often contributes to the enhanced expression of inflammatory genes, such as genes of inflammatory cytokines, during pathogen and TLR ligand stimulation (36, 37). MKP-1 is known to regulate the mRNA stability of numerous inflammatory genes (38, 39, 40). We investigated whether Mkp-1 deficiency alters the half-life of Pfkfb3 mRNA in macrophages stimulated by LPS. To induce Pfkfb3 mRNA expression, we stimulated both WT and Mkp-1−/− BMDM with LPS. Gene transcription was then paused using actinomycin D, and time-dependent decay of Pfkfb3 mRNA was assessed using qRT-PCR (Fig. 5D). Pfkfb3 mRNA decayed gradually with somewhat similar rates in WT and Mkp-1–deficient BMDM after actinomycin D treatment, with an estimated half-life of 2.84 and 2.33 h, respectively. Since Mkp-1 deficiency did not enhance the stability of Pfkfb3 mRNA, enhanced Pfkfb3 expression in Mkp-1−/− BMDM was likely caused by enhanced Pfkfb3 transcription.
Pfkfb3 plays an important role in enhancing glycolysis in LPS-stimulated macrophages
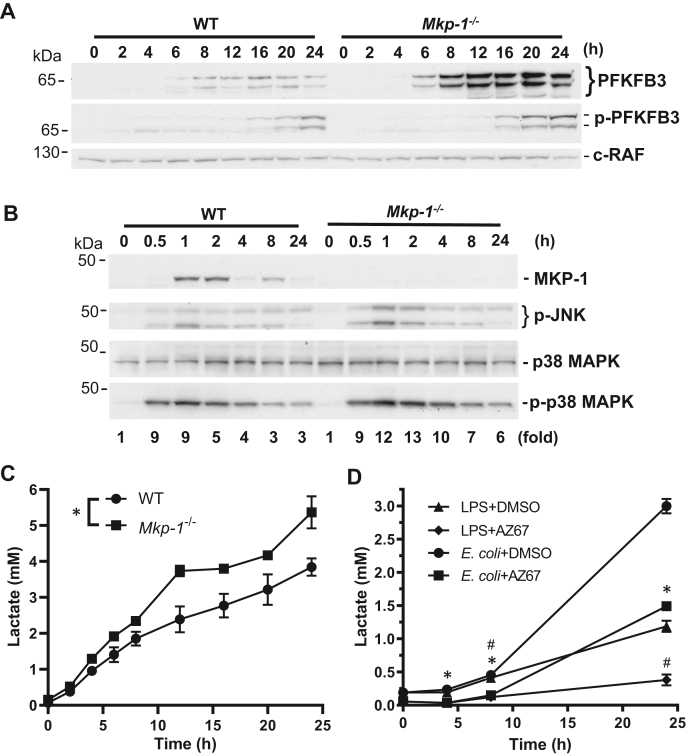
To understand the role of PFKFB3 in glycolysis in macrophages after inflammatory stimulation, we assessed the kinetics of PFKFB3 expression and phosphorylation as well as the lactate production in both WT and Mkp-1−/− macrophages (Fig. 6). Without LPS stimulation, PFKFB3 levels were nearly undetectable in both WT and Mkp-1−/− BMDM (Fig. 6A). Upon LPS stimulation, PFKFB3 protein gradually increased; it was clearly detectable by 6 h and continued to increase over time in both WT and Mkp-1−/− cells, reaching its plateau after 8 to 12 h. Compared to WT cells, PFKFB3 protein levels were substantially greater in Mkp-1−/− cells, particularly for the 62 and 70 kDa isoforms. Similarly, LPS stimulation triggered a gradual increase in the levels of S461-phosphorylated PFKFB3 in both WT and Mkp-1−/− macrophages. Unlike PFKFB3, which reached plateau after 8 to 12 h, Ser461-phosphorylated PFKFB3 continued to increase up to 24 h. Phospho-PFKFB3 levels also appeared higher in Mkp-1−/− macrophages than in WT macrophages. MKP-1 protein was induced by LPS in WT but not in Mkp-1−/− BMDM, and MKP-1 expression declined with time in WT BMDM (Fig. 6B). The levels of both phospho-p38 and phospho-JNK MAPKs were substantially increased in response to LPS stimulation in both WT and Mkp-1−/− BMDM (Fig. 6B) and sustained longer in LPS-stimulated Mkp-1−/− BMDM than in LPS-stimulated WT BMDM (Fig. 6B). These results are consistent with the results published in previous studies (31, 41, 42, 43, 44), To assess lactate production after LPS stimulation in WT and Mkp-1−/− BMDM, cells were fed fresh pyruvate-free medium and then stimulated with LPS. The medium was harvested at different times following LPS stimulation. Upon LPS stimulation, lactate levels gradually increased in the culture medium of both WT and Mkp-1−/− BMDM. Compared to WT BMDM, Mkp-1−/− BMDM produced significantly greater amounts of lactate (Fig. 6C). To determine the contribution of PFKFB3 to the regulation of glycolysis in macrophages, WT BMDM were fed fresh pyruvate-free medium and then pretreated with a pharmacological inhibitor highly selective for PFKFB3, AZ67 (45, 46), or vehicle DMSO. Thirty minutes later, these cells were treated with either heat-killed E. coli at the ratio of 10 E. coli cells per macrophage or LPS at a concentration of 100 ng/ml. Medium was collected at different times and lactate levels were assayed (Fig. 6D). Both LPS and E. coli induced a marked increase in lactate production at 8 and 24 h, although E. coli led to a more robust lactate production. AZ67 almost abolished lactate production induced by LPS and substantially attenuated lactate production induced by E. coli. Similar inhibition of lactate production by AZ67 was also seen in Mkp-1−/− BMDM (data not shown). These results clearly indicate a critical role of PFKFB3 in the regulation of glycolysis in macrophages during the inflammatory response.
Figure 6.
Enhanced PFKFB3 expression in Mkp-1−/−macrophages is associated with accelerated glycolysis.A, the kinetics of PFKFB3 induction and phosphorylation following LPS stimulation in WT and Mkp-1−/− BMDM. WT and Mkp-1−/− BMDM were cultured in medium overnight and then stimulated with 100 ng/ml LPS. Cells were lysed at different times. PFKFB3 expression and phosphorylation were assessed by Western blot analyses using PFKFB3 (Upper panel) or phospho-PFKFB3 (Middle panel) antibody. c-RAF antibody was used to control for protein loading (Lower panel). B, kinetics of MKP-1 expression, JNK, and p38 MAPK activation in BMDM after LPS stimulation. WT and Mkp-1−/− BMDM were cultured overnight and then stimulated with 100 ng/ml LPS. Cells were lysed at different times for Western blotting with an MKP-1, phosphor-JNK, phosphor-, or total p38 MAPK antibody. The ratios between phosphor-p38 MAPK and total p38 MAPK were calculated and presented as fold of changes relative to the controls (time 0) underneath the p-p38 MAPK blot. Representative Western blotting results are shown. C, kinetics of lactate production following LPS stimulation. WT and Mkp-1−/− BMDM were cultured in pyruvate-free medium containing 10% dialyzed FBS overnight. The next day, cells were fed with fresh medium and stimulated with LPS. Lactate concentration in the medium was assayed using a fluorometric lactate assay kit. Values were presented as means ± SE. ∗p < 0.05, compared to WT BMDM (two-way ANOVA, n = 3–4). There was an interaction between genotype and time. D, pharmacological inhibition of PFKFB3 attenuates lactate production by LPS- or E. coli–stimulated macrophages. WT BMDM grown overnight were first fed with fresh medium and then were pretreated with AZ67 or DMSO (vehicle) for 30 min. The cells were stimulated with heat-killed E. coli (MOI: 10:1) or 100 ng/ml LPS for different times. Medium was harvested and lactate concentrations in the medium were measured using a fluorometric lactate assay kit. ∗p < 0.05, compared to DMSO-pretreated, E. coli-stimulated cells; #, p < 0.05, compared to DMSO-pretreated, LPS-stimulated cells (two-way ANOVA, n = 3). BMDM, bone marrow–derived macrophage; LPS, lipopolysaccharide; MAPK, mitogen-activated protein kinase; MKP, MAPK phosphatase; PFKFB, phosphofructo-2-kinase/fructose-2,6-biphosphatase.
In macrophages, both PFKFB3 induction and enhanced glycolysis following LPS stimulation are primarily mediated by p38 MAPK
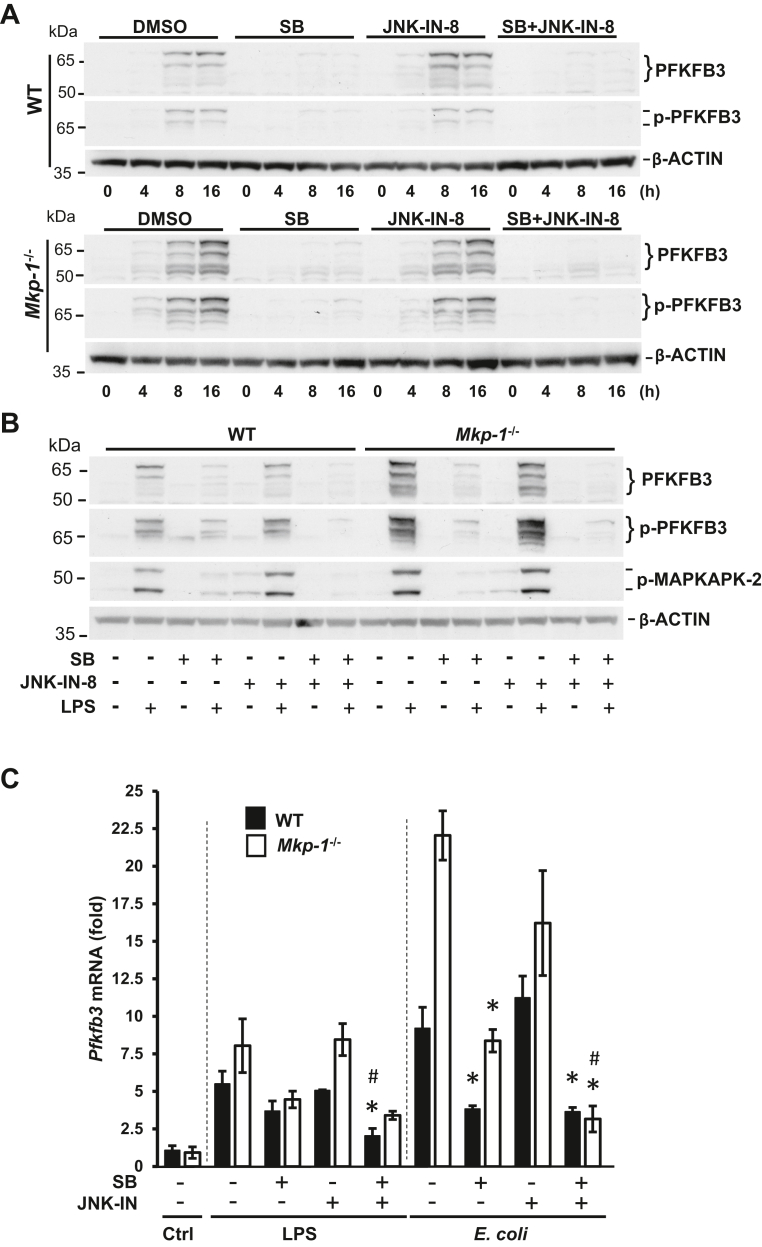
MKP-1 is a negative regulator of p38 and JNK MAPKs, and in macrophages, Mkp-1 deficiency leads to prolonged activation of p38 and JNK MAPKs (31, 47). To understand the role of p38 and JNK MAPKs in the regulation of PFKFB3 and glycolysis, we pretreated WT and Mkp-1−/− BMDM with a pharmacological inhibitor of p38 MAPK (SB203580) or JNK (JNK-IN-8) alone or with both inhibitors. After 15 min, we stimulated cells with LPS for 4, 8, and 16 h and then assessed the effects of these inhibitors on PFKFB3 protein (Fig. 7A). In WT BMDM, LPS stimulated a modest increase in total and Ser461-phosphorylated PFKFB3. In Mkp-1−/− BMDM, LPS stimulated PFKFB3 expression more robustly, especially the 62 and 70 kDa isoforms. The p38 MAPK inhibitor substantially inhibited both PFKFB3 induction and PFKFB3 phosphorylation in both WT and Mkp-1−/− BMDM, while the JNK inhibitor alone had little effect on either the induction or the phosphorylation of PFKFB3. Combining the p38 MAPK and JNK inhibitors nearly abolished the induction of the total and phosphorylated PFKFB3 proteins. Side-by-side comparison of the effects of the p38 MAPK and JNK inhibitors after 16 h LPS stimulation largely confirmed these observations (Fig. 7B). The p38 MAPK inhibitor had a more potent inhibitory effect on both total and Ser461-phosphorylated PFKFB3 proteins, while the JNK inhibitor had little inhibitory effect (Fig. 7B). Combination of the two inhibitors attenuated total and phosphorylated PFKFB3 proteins in Mkp-1−/− BMDM to levels comparable to those in WT BMDM. Supporting the substantial inhibition of p38 by SB203580, MAPKAPK-2 phosphorylation induced by LPS was substantially attenuated by SB203580, regardless of being used by itself or in combination with JNK-IN-8, in both WT and Mkp-1−/− BMDM (Fig. 7B). These results indicate that enhanced PFKFB3 induction and phosphorylation in the Mkp-1−/− BMDM is primarily due to the enhanced p38 MAPK activity, and JNK plays a minor role in PFKFB3 regulation.
Figure 7.
PFKFB3 induction in macrophages following inflammatory stimuli is primarily mediated by p38 MAPK.A, the effect of p38 MAPK and JNK inhibition on PFKFB3 induction and phosphorylation after LPS stimulation in WT and Mkp-1−/− BMDM. BMDM were treated with DMSO, 10 μM SB203580, 3 μM JNK-IN-8, or 10 μM SB203580 plus 3 μM JNK-IN-8 for 15 min and then stimulated with LPS for 0, 4, 8, or 16 h. Cell lysates were separated on duplicate gels to detect total and Ser461-phosphorylated PFKFB3, respectively, by Western blot analysis. The membranes were stripped and blotted with a mouse β-actin antibody to verify comparable sample loading. Note: the blots for WT and Mkp-1−/− samples were processed together and exposed to the same film (images of WT and Mkp-1−/− samples from the same film were used). B, p38 MAPK plays a primary role in the induction of PFKFB3. WT and Mkp-1−/− BMDM were treated with DMSO, 10 μM SB203580, 3 μM JNK-IN-8, or 10 μM SB203580 plus 3 μM JNK-IN-8 for 15 min and then stimulated without or with LPS for 16 h. PFKFB3, phosphor-PFKFB3, and phosphor-MAPKAPK-2 levels were assessed by Western blotting. Representative results were shown. C, the effects of p38 MAPK and JNK inhibitors on Pfkfb3 mRNA levels following LPS and Escherichia coli stimulation. WT and Mkp-1−/− BMDM were pretreated with DMSO, 10 μM SB203580, 3 μM JNK-IN-8, or 10 μM SB203580 plus 3 μM JNK-IN-8 for 15 min and then stimulated without or with either heat-killed E. coli (MOI: 10:1) or 100 ng/ml LPS for 8 h. Pfkfb3 mRNA levels in the samples were quantitated by qRT-PCR. The results were normalized to 18S ribosomal RNA. The expression of mRNA is presented as fold change relative to WT control cells pretreated with DMSO. ∗p < 0.05, compared to LPS- or E. coli–stimulated cells of the same genotype; #, p < 0.05, compared to SB203580-pretreated, LPS- or E. coli–stimulated cells of the same genotype (t test, n = 3–6). BMDM, bone marrow–derived macrophage; LPS, lipopolysaccharide; MAPK, mitogen-activated protein kinase; MKP, MAPK phosphatase; MAPKAPK, MAPK-activated protein kinase; PFKFB, phosphofructo-2-kinase/fructose-2,6-biphosphatase; qRT-PCR, quantitative RT-PCR.
The effects of p38 MAPK and JNK inhibition on Pfkfb3 mRNA expression in BMDM stimulated with LPS and E. coli were assessed via qRT-PCR. In the presence of vehicle (DMSO), LPS induced 5.5- and 8.4-fold increases by 8 h in WT and Mkp-1−/− BMDM, respectively. E. coli induced 8.5- and 22-fold increases in Pfkfb3 mRNA by 8 h in WT and Mkp-1−/− BMDM, respectively (Fig. 7C). The p38 MAPK inhibitor attenuated Pfkfb3 induction by E. coli, while it also appeared to dampen Pfkfb3 induction by LPS. In contrast, the JNK inhibitor had little effect in either WT or Mkp-1−/− BMDM. The inhibition of p38 MAPK and JNK together appeared to exert a greater inhibition on Pfkfb3 induction, particularly in E. coli–stimulated Mkp-1−/− BMDM. In fact, when both p38 MAPK and JNK were inhibited, E. coli–induced Pfkfb3 expression was similar in WT and Mkp-1−/− BMDM.
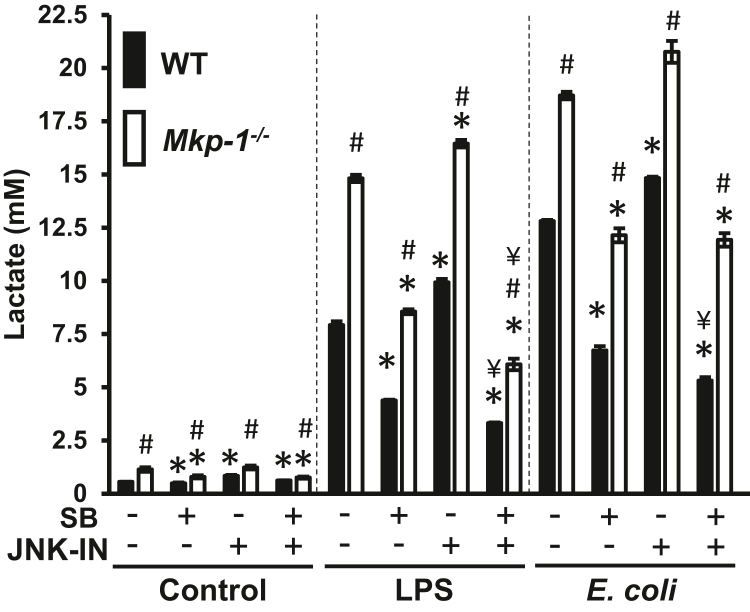
The effects of p38 MAPK and JNK inhibition on glycolysis were examined (Fig. 8). Without LPS stimulation, lactate production is very low, regardless of whether BMDM were treated with p38 MAPK or JNK inhibitors. LPS stimulation substantially enhanced lactate production. Compared to BMDM treated by LPS, BMDM derived from both WT and Mkp-1 KO mice produced more lactate after stimulation with heat-killed E. coli. The p38 MAPK inhibitor significantly suppressed lactate production in both LPS- and E. coli–stimulated BMDM, regardless of whether they originated from WT or Mkp-1 KO mice. Surprisingly, the JNK inhibitor slightly, yet significantly, enhanced lactate production in both WT and Mkp-1−/− BMDM following either LPS or E. coli stimulation. Combining the p38 MAPK and JNK inhibitors further blocked lactate production in both WT and Mkp-1−/− BMDM stimulated by LPS. While the combination of both the p38 MAPK and JNK inhibitors had a greater inhibition on lactate production in E. coli–stimulated WT BMDM compared to the p38 MAPK inhibitor alone, the combination of the two inhibitors did not further attenuate lactate production in E. coli–stimulated Mkp-1−/− BMDM in relation to the p38 MAPK inhibitor alone. Taken together, these results support a critical role of p38 MAPK in the regulation of PFKFB3 expression and glycolysis during the inflammatory response.
Figure 8.
Glycolysis in macrophages following inflammatory stimuli is primarily mediated by p38 MAPK. WT and Mkp-1−/− BMDM were plated into a 24-well plate. The next day, cells were fed with fresh medium treated with DMSO, 10 μM SB203580, 3 μM JNK-IN-8, or 10 μM SB203580 plus 3 μM JNK-IN-8 for 30 min and then stimulated with 100 ng/ml LPS or heat-killed Escherichia coli (MOI: 10:1) for 16 h or left unstimulated. Medium was harvested and lactate levels in the media were measured using a fluorometric lactate assay kit. ∗p < 0.05, compared to DMSO-pretreated samples of the same genotype under the same stimulation; #, p < 0.05, compared to WT samples of the same stimulation and pretreatment. ¥, <0.05, compared to samples of the same genotype pretreated with SB203580 and of same treatment (t test, n = 4). MAPK, mitogen-activated protein kinase; MKP, MAPK phosphatase; BMDM, bone marrow–derived macrophage; LPS, lipopolysaccharide.
Discussion
Although sepsis is known to enhance glycolysis, the mechanisms involved are not fully understood. In this study, we demonstrated that E. coli–induced sepsis not only strongly increased the expression of PFKFB3 as a nuclear protein in a variety of tissues (Figure 1, Figure 2, Figure 3) but also enhanced the levels of Ser461-phosphorylated PFKFB3 (Figure 1, Figure 2, Figure 3). The immunohistochemical studies demonstrate that 1) PFKFB3 is expressed preferentially in certain cell types and 2) PFKFB3 expression is increased upon infection with E. coli. Following infection, PFKFB3 was most prominently expressed in hepatocytes, renal proximal tubules, collecting ducts, vascular endothelium, circulating leukocytes including monocytes, and tissue-resident macrophages across organ systems (Figs. 4 and S2–S5). Moreover, sepsis-induced PFKFB3 expression was strongly potentiated in the absence of a functional Mkp-1 gene (Figure 3, Figure 4 and S2–S5). Using BMDM, we studied the regulation of PFKFB3 and its function in glycolysis during inflammatory response. We found that enhanced PFKFB3 expression was associated with increased Pfkfb3 mRNA expression (Fig. 5). Additionally, Mkp-1 deficiency augmented Pfkfb3 mRNA expression through a mechanism primarily mediated by p38 MAPK (Fig. 7C) without increasing the stability of Pfkfb3 mRNA (Fig. 5C). We found that LPS- and E. coli–enhanced lactate production coincided with PFKFB3 induction and phosphorylation, and it was markedly attenuated by a selective PFKFB3 inhibitor (Fig. 6). Pharmacological inhibition of p38 MAPK markedly decreased PFKFB3 expression and Ser461-phosphorylated PFKFB3 protein levels (Fig. 7) and substantially attenuated lactate production (Fig. 8). These results clearly demonstrate the critical role of PFKFB3 in glycolysis and further highlight the role of p38 MAPK in regulating this process. These studies suggest that glycolytic reprogramming following pathogen infection in sepsis patients is organized by the p38 MAPK pathway and, as a critical negative regulator of p38 MAPK, MKP-1 participates in the regulation of the glycolytic process.
The mechanism by which Mkp-1 deficiency enhances glycolysis
We have previously shown that Mkp-1 KO mice exhibit an enhanced inflammatory response, increased mortality, and a substantially altered liver metabolic program relative to WT mice following both LPS and E. coli challenges (31, 48, 49). Furthermore, we have found that Mkp-1−/− macrophages produce elevated cytokines in response to inflammatory stimuli. In this study, we found that Mkp-1 deficiency enhanced the mRNA expression of several rate-limiting glycolytic enzymes in the livers of mice infected with E. coli, including HK1, HK2, PFKFB3, and PFKP (Fig. 1A). Among these, the substantial elevation of PFKFB3 in response to sepsis and its regulation by MKP-1 are particularly interesting for several reasons. First, PFKFB3 has the highest kinase-to-phosphatase activity ratio (20). Enhanced PFKFB3 activity favors the accumulation of F-2,6-bisP, a potent allosteric activator of the rate-limiting enzyme of glycolysis, PFK1. For this reason, increases in PFKFB3 activity should dramatically stimulate the glycolytic cascade. Second, following inflammatory stimulation, Pfkfb3 was the most highly induced glycolysis-related gene in the livers of Mkp-1 KO mice (Fig. 1) and Mkp-1−/− macrophages (data not shown). The dramatically increased Pfkfb3 mRNA in LPS-stimulated Mkp-1−/− BMDM (Fig. 5, A and B) and the potent inhibitory effect of SB203580 on Pfkfb3 mRNA induction (Fig. 7C) strongly support enhanced p38 MAPK activity as a strong positive regulator of Pfkfb3 transcription. The substantial inhibition of lactate production in LPS- and E. coli–stimulated BMDM (Fig. 8) clearly demonstrates the critical role of the p38 MAPK pathway and MKP-1–mediated negative feedback in the regulation of glycolysis. Third, at least in the liver, the PFKFB3 isoforms expressed in WT and Mkp-1 KO mice appeared to differ; compared to E. coli–infected WT mice that had 61 and 67 kDa as their major isoforms, Mkp-1 KO mice preferentially express different PFKFB3 isoforms (Fig. 1, C and D). Mice express eight PFKFB3 isoforms, due to differential mRNA splicing (50, 51), however it remains unclear whether the PFKFB3 isoforms of varying weights function differently during sepsis. The differential utilization of the distinct isoforms in WT and Mkp-1 KO mice following sepsis suggests that the splicing of the Pfkfb3 mRNA is affected directly or indirectly by Mkp-1 deficiency. Last, PFKFB3 activity is regulated not only by protein expression but also by phosphorylation (19, 20). It has been shown that a variety of protein kinases can phosphorylate PFKFB3, including PKA, PKC, MAPKAPK-2, AMPK, and CDK6 (19, 20, 21, 22), leading to increased catalytic activity. MAPKAPK-2 is a direct target of p38 MAPK, and phosphorylation of MAPKAPK-2 by p38 MAPK enhances its kinase activity (29, 52). Mkp-1 KO mice exhibited greater phospho-PFKFB3/PFKFB3 ratios than WT mice, at least in the lungs and kidneys (data not shown), suggesting that these organs likely have augmented PFKFB3 activity. Thus, elevated p38 MAPK activity in these organs (Fig. 2, B and C) provides a plausible explanation for increased PFKFB3 phosphorylation at Ser461 in these organs in the E. coli–infected Mkp-1 KO mice. As increased lactate during sepsis has been implicated in immune suppression and vascular leak (14, 53, 54), enhanced glycolysis may contribute to the elevated bacterial burden, vascular collapse, and increased mortality of Mkp-1 KO mice following E. coli infection (48, 55).
While Mkp-1 deficiency enhances the expression of PFKFB3 during the inflammatory response, the mechanism involved remains unclear. Because Mkp-1 deficiency enhanced Pfkfb3 mRNA expression without increasing the mRNA stability (Fig. 5D), it is likely that Mkp-1 deficiency results in augmented Pfkfb3 transcription. Since the p38 MAPK inhibitor significantly attenuated Pfkfb3 mRNA induction, particularly in Mkp-1−/− BMDM following E. coli stimulation (Fig. 7C), enhanced p38 MAPK activity in Mkp-1−/− cells is probably responsible for enhanced Pfkfb3 transcription. Previously, Rius et al. have demonstrated that the hypoxic response in innate immunity is mediated by HIF1α and NF-κB (56, 57). Obach et al. have shown that the human PFKFB3 gene promoter contains several putative HIF1α-binding sites necessary for transactivation in response to hypoxia (26). Kwon et al. have shown that p38 MAPK can phosphorylate HIF1α and that such phosphorylation favors HIF1α accumulation (58). Thus, in theory, enhanced p38 MAPK activity as the result of Mkp-1 deficiency may lead to greater HIF1α levels and enhancement in HIF1α-mediated gene transcription. However, following LPS stimulation, WT and Mkp-1−/− BMDM had similar HIF1α levels (data not shown). We have previously shown that Mkp-1 deficiency does not have an obvious effect on NF-κB pathway activation in LPS-stimulated BMDM (40). Thus, enhanced Pfkfb3 expression in Mkp-1−/− macrophages following LPS and E. coli stimulation is probably mediated via a p38 MAPK-regulated transcriptional factor(s), rather than NF-κB or HIF1α.
Mkp-1 deficiency enhances both p38 MAPK and JNK activities in LPS-stimulated macrophages (31, 41, 42, 43). Although it is abundantly clear that p38 MAPK regulates PFKFB3 activity by enhancing its expression and phosphorylation, the role of JNK in PFKFB3 and glycolysis regulation remains puzzling. The JNK inhibitor by itself had little effect on PFKFB3 protein expression (Fig. 7, A and B) in BMDM stimulated by LPS, whereas the combination of the p38 MAPK and JNK inhibitors further decreased PFKFB3 protein and Pfkfb3 mRNA expression in LPS- and E. coli–stimulated macrophages relative to the p38 MAPK inhibitor alone (Fig. 7). These results suggest that JNK plays a minor role in the induction of PFKFB3. Surprisingly, pharmacological inhibition of JNK alone resulted in a small, yet significant, enhancement of lactate production (Fig. 8). One possibility is that JNK inhibition could enhance the activity of one or more key glycolytic enzymes that function(s) in the glycolytic pathway. This is consistent with the report that knockdown of JNK1 in normal mouse liver cells upregulates the hepatic expression of clusters of genes involved in glycolysis, including Hk2, Gpi1, and Pkm (59). Additionally, JNK inhibition has been shown to increase HK2 enzymatic activity in Chaetocin-treated glioma cells (60). Consistent with this observation, we also found that Mkp-1−/− BMDM express less HK2 protein than WT BMDM both before and after LPS stimulation (data not shown). It is possible that when p38 MAPK and JNK are both inhibited, PFKFB3 activity is markedly attenuated, making PFK1 activity the bottle-neck of the glycolytic cascade that constrains the rate of lactate production, explaining the greater inhibition of lactate production than the p38 MAPK inhibitor alone.
Experimental procedures
Experimental animals
Mkp-1+/− mice on a C57/129 mixed background (61, 62) were generously provided by Bristol-Myers Squibb Pharmaceutical Research Institute. Mkp-1+/− mice were intercrossed to generate WT and Mkp-1 KO mice for E. coli infection experiments. Mkp-1 KO mice have no obvious phenotype prior to E. coli infection. Additionally, the Mkp-1+/− mice were backcrossed to C57BL/6J mice for eight generations to create Mkp-1 KO mice on a C57BL/6J background. While WT and Mkp-1 KO mice on C57/129 background were used for all infection experiments, all macrophage studies in vitro were carried out using bone marrow isolated from the mice on C57BL/6J background. All mice were housed with a 12 h alternating light-dark cycle at 25 °C, with humidity between 30% and 70%, and have access to food and water ad libitum. All experiments were performed according to National Institutes of Health guidelines and were approved by the Institutional Animal Care and Use Committee at the Research Institute at Nationwide Children’s Hospital.
E. coli infection and RNA-seq
A WT (smooth) strain of E. coli (O55:B5, ATCC 12014) was purchased from American Tissue Culture Collection. E. coli were grown, prepared, and used to infect mice via tail vein injection at a dose of 2.5 × 107 CFU/g body weight (b.w.) as previously described (27, 48). Livers, lungs, spleens, kidneys, and hearts were isolated 24 h postinfection. Tissues were either fixed in formalin for IHC or freeze-clamped and stored at −80 °C. Tissues were homogenized to extract proteins for Western blot analysis. Total RNA was isolated from four animals in each treatment group for RNA-seq analysis (27, 40, 49). The RNA-seq data have been deposited in Gene Expression Omnibus (GSE122741). A comprehensive list of 23 glycolysis-related genes was compiled, and the transcript copy numbers were used to calculate the fold change and p-values using a t test. The fold change of transcripts for each gene was calculated relative to the average expression in control WT mice (injected with PBS, i.v.). Values were log2-transformed to generate a heatmap where red indicates upregulation, white indicates no change, and blue indicates downregulation of gene expression.
Macrophage derivation and stimulation
BMDMs were generated from WT and Mkp-1 KO mice on the C57BL/6J background as previously described (40) and were stimulated with LPS (O55:B5, Calbiochem) or heat-killed E. coli for different times. In some experiments, BMDM were pretreated with vehicle (DMSO), a p38 MAPK inhibitor (SB203580 (63), Calbiochem), a JNK inhibitor (JNK-IN-8 (64), Selleck Chemicals), or a combination of both inhibitors for 15 min prior to LPS stimulation. Medium was harvested to measure lactate concentrations using a Lactate-Glo assay kit (Promega). Cells were lysed to harvest proteins for Western blot analysis.
Quantitative RT-PCR
Total RNA was isolated either from liver tissues or BMDM using Trizol. RQ1 RNase-Free DNase (Promega) was used to remove Genomic DNA from total RNA samples prior to reverse transcription, as previously described (27, 40). Pfkfb3 mRNA levels were assessed by qRT-PCR using forward primer 5′-AGAACTTCCACTCTCCCACCC-3′ and reverse primer 5′-AGGGTAGTGCCCATTGTTGAA-3’. For an internal control for normalization, 18S rRNA was quantified by qRT-PCR using primers 5′-GTAACCCGTTGAACCCCATT-3′ and 5′-CCATCCAATCGGTAGTAGCG-3’. Pfkfb3 mRNA expression was normalized to 18S using the 2-ΔΔCT method (36). The expression of Pfkfb3 mRNA in liver tissues was also assessed similarly by qRT-PCR (27).
Assessment of Pfkfb3 mRNA stability
To assess the effect of Mkp-1 deficiency on Pfkfb3 mRNA half-life, WT and Mkp-1−/− BMDM were stimulated with LPS (100 ng/ml) for 8 h. Gene transcription was then stopped by 5 μg/ml actinomycin D, as previously described (65). RNA samples were isolated after different times, and Pfkfb3 mRNA levels were assessed by qRT-PCR. The half-life of Pfkfb3 mRNA was calculated using the formula, where and and is the half-life.
Western blot analysis and IHC
Western blot analysis was done as described previously (66, 67). The rabbit polyclonal antibody against PFKFB3 was purchased from Proteintech. The rabbit polyclonal antibody against phospho-PFKFB3 (Ser461) was purchased from Thermo Fisher Scientific. The mouse mAb against β-ACTIN was purchased from Sigma Chemicals. The mouse mAb against c-RAF was purchased from Transduction Laboratories. The rabbit mAbs against GAPDH, MKP-1, Thr180/Tyr182-phosphorylated and total p38, and the polyclonal rabbit antibody against Thr334-phosphorylated MAPKAPK-2 were purchased from Cell Signaling Technology. Western blots were developed using chemiluminescent reagent ECL Immobilon (Millipore Corporation). Western blot images were acquired using the Epson Perfection 4990 PHOTO scanner (Epson). Quantification of protein expression was carried out by densitometry using VisionWorksLS Image Acquisition and Analysis Software (UVP), as previously described (66).
IHC was carried out as previously described (68). Briefly, 5-μm paraffin tissue sections were deparaffinized in xylene and rehydrated with graded ethanol to potassium-PBS solution, pH 7.2. After antigen retrieval with citrate buffer (pH 6), the sections were pretreated with 1.5% H2O2 for 15 min, followed by 1 h blocking with 5% normal donkey serum (Jackson ImmunoResearch). The tissues were then incubated overnight at 4 °C with the rabbit polyclonal antibody against PFKFB3 diluted 1:8000 in 5% normal donkey serum. After 1 h incubation with biotinylated donkey anti-rabbit IgG 1:600 dilution (Jackson ImmunoResearch), the sections were developed using the avidin-biotin-peroxidase system (Vectastain Elite ABC; Vector Laboratories) with Vector NovaRed (Vector Laboratories) as chromogen and hematoxylin as counterstain. The specificity of the immunoreactivity was confirmed by omission of the PFKFB3 antibody. To quantify the PFKFB3 staining in hepatocyte nuclei, we used a semiquantitative histoscore system based on the intensity of staining graded as follows: 0, nonstaining; 1, weak; 2, moderate; or 3, strong, according to a previously described semiquantitative scoring system (69). Four randomly obtained representative fields, including centrilobular, midzonal, and portal regions (each field covering 0.03 mm2), were scored. The mean hepatocyte histoscore was calculated for each field, from a total of 50 to 100 cells.
Lactate assays
BMDM were plated into 96-well plates at a density of 2.5 × 105 cells per well. After the cells attached, the medium was removed and cells were washed twice with PBS. The cells were fed with fresh pyruvate-free medium containing dialyzed FBS and stimulated with LPS or heat-killed E. coli. When the pharmacological PFKFB3 inhibitor, AZ67, was used, cells were pretreated with 10 μM AZ67 or vehicle (DMSO) for 30 min and then stimulated with LPS or E. coli. Medium was harvested after different times and lactate levels in the medium were measured using a fluorometric lactate assay kit (Promega).
Statistical analyses
Differences in protein and gene expression between groups were compared using t test or two-way ANOVA with GraphPad Prism 8.2.0 program (GraphPad Software). A value of p < 0.05 was considered statistically significant for all analyses.
Data availability
The RNA-seq data have been deposited in the Gene Expression Omnibus (GSE122741) https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE122741.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We are grateful to Bristol-Myers Squibb Pharmaceutical Research Institute for providing Mkp-1 KO mice. We thank Drs. Xianxi Wang, Jinhui Li, Dimitrios Anastasakis, and William E. Ackerman IV for their contributions to in vivo infection and RNA-seq experiments. We also gratefully acknowledge Cynthia McAllister for technical assistance.
Author contributions
C. E. M., P. R. M., T, J. B., X. W., K. L., and Y. L. methodology; C. E. M., J. M. M., E. D. S., P. R. M., B. A. B., T. J. B., X. W., S. C. L., and K. L. investigation; C. E. M., J. M. M., E. D. S., P. R. M., B. A. B., T. J. B., and X. W. data curation; C. E. M., B. A. B., S. C. L., L. D. N., and Y. L. writing–review and editing; E. D. S., L. D. N., and Y. L. formal analysis; L. D. N. and Y. L. funding acquisition; M. H. resources; M. H. and Y. L. supervision; Y. L. conceptualization; Y. L. project administration; Y. L. writing–original draft.
Funding and additional information
This work was supported by grants from NIH (AI124029 and AI142885 to Y. L.). S. C. L. is supported by a Genentech Veterinary Pathology Fellowship through Genentech Inc and The Ohio State University. The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Reviewed by members of the JBC Editorial Board. Edited by Wolfgang Peti
Supporting information
References
- 1.Singer M., Deutschman C.S., Seymour C.W., Shankar-Hari M., Annane D., Bauer M., et al. The third international consensus definitions for sepsis and septic shock (Sepsis-3) Jama. 2016;315:801–810. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shankar-Hari M., Phillips G.S., Levy M.L., Seymour C.W., Liu V.X., Deutschman C.S., et al. Developing a new definition and assessing new clinical criteria for septic shock: for the third international consensus definitions for sepsis and septic shock (Sepsis-3) JAMA. 2016;315:775–787. doi: 10.1001/jama.2016.0289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shapiro N.I., Howell M.D., Talmor D., Nathanson L.A., Lisbon A., Wolfe R.E., et al. Serum lactate as a predictor of mortality in emergency department patients with infection. Ann. Emerg. Med. 2005;45:524–528. doi: 10.1016/j.annemergmed.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 4.Noritomi D.T., Soriano F.G., Kellum J.A., Cappi S.B., Biselli P.J., Libório A.B., et al. Metabolic acidosis in patients with severe sepsis and septic shock: a longitudinal quantitative study. Crit. Care Med. 2009;37:2733–2739. doi: 10.1097/ccm.0b013e3181a59165. [DOI] [PubMed] [Google Scholar]
- 5.Suetrong B., Walley K.R. Lactic acidosis in sepsis: it's not all anaerobic: implications for diagnosis and management. Chest. 2016;149:252–261. doi: 10.1378/chest.15-1703. [DOI] [PubMed] [Google Scholar]
- 6.Boekstegers P., Weidenhöfer S., Kapsner T., Werdan K. Skeletal muscle partial pressure of oxygen in patients with sepsis. Crit. Care Med. 1994;22:640–650. doi: 10.1097/00003246-199404000-00021. [DOI] [PubMed] [Google Scholar]
- 7.Haupt M.T., Gilbert E.M., Carlson R.W. Fluid loading increases oxygen consumption in septic patients with lactic acidosis. Am. Rev. Respir. Dis. 1985;131:912–916. doi: 10.1164/arrd.1985.131.6.912. [DOI] [PubMed] [Google Scholar]
- 8.Sair M., Etherington P.J., Peter Winlove C., Evans T.W. Tissue oxygenation and perfusion in patients with systemic sepsis. Crit. Care Med. 2001;29:1343–1349. doi: 10.1097/00003246-200107000-00008. [DOI] [PubMed] [Google Scholar]
- 9.VanderMeer T.J., Wang H., Fink M.P. Endotoxemia causes ileal mucosal acidosis in the absence of mucosal hypoxia in a normodynamic porcine model of septic shock. Crit. Care Med. 1995;23:1217–1226. doi: 10.1097/00003246-199507000-00011. [DOI] [PubMed] [Google Scholar]
- 10.Opdam H., Bellomo R. Oxygen consumption and lactate release by the lung after cardiopulmonary bypass and during septic shock. Crit. Care Resusc. 2000;2:181–187. [PubMed] [Google Scholar]
- 11.Gore D.C., Jahoor F., Hibbert J.M., DeMaria E.J. Lactic acidosis during sepsis is related to increased pyruvate production, not deficits in tissue oxygen availability. Ann. Surg. 1996;224:97–102. doi: 10.1097/00000658-199607000-00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bar-Or D., Carrick M., Tanner A., 2nd, Lieser M.J., Rael L.T., Brody E. Overcoming the Warburg effect: is it the key to survival in sepsis? J. Crit. Care. 2018;43:197–201. doi: 10.1016/j.jcrc.2017.09.012. [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Alvarez M., Marik P., Bellomo R. Sepsis-associated hyperlactatemia. Crit. Care. 2014;18:503. doi: 10.1186/s13054-014-0503-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nolt B., Tu F., Wang X., Ha T., Winter R., Williams D.L., et al. Lactate and immunosuppression in sepsis. Shock. 2018;49:120–125. doi: 10.1097/SHK.0000000000000958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wasyluk W., Zwolak A. Metabolic alterations in sepsis. J. Clin. Med. 2021;10 doi: 10.3390/jcm10112412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O'Neill L.A., Pearce E.J. Immunometabolism governs dendritic cell and macrophage function. J. Exp. Med. 2016;213:15–23. doi: 10.1084/jem.20151570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chou W.C., Rampanelli E., Li X., Ting J.P. Impact of intracellular innate immune receptors on immunometabolism. Cell Mol. Immunol. 2022;19:337–351. doi: 10.1038/s41423-021-00780-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nelson D.L.C., M M., Hoskins A.A. Eighth Edition. W.H. Freeman and Company; New York, NY: 2021. Lehninger Principles of Biochemistry. [Google Scholar]
- 19.Yi M., Ban Y., Tan Y., Xiong W., Li G., Xiang B. 6-Phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 and 4: a pair of valves for fine-tuning of glucose metabolism in human cancer. Mol. Metab. 2019;20:1–13. doi: 10.1016/j.molmet.2018.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sakakibara R., Kato M., Okamura N., Nakagawa T., Komada Y., Tominaga N., et al. Characterization of a human placental fructose-6-phosphate, 2-kinase/fructose-2,6-bisphosphatase. J. Biochem. 1997;122:122–128. doi: 10.1093/oxfordjournals.jbchem.a021719. [DOI] [PubMed] [Google Scholar]
- 21.Bolanos J.P. Adapting glycolysis to cancer cell proliferation: the MAPK pathway focuses on PFKFB3. Biochem. J. 2013;452:e7–9. doi: 10.1042/BJ20130560. [DOI] [PubMed] [Google Scholar]
- 22.Novellasdemunt L., Bultot L., Manzano A., Ventura F., Rosa J.L., Vertommen D., et al. PFKFB3 activation in cancer cells by the p38/MK2 pathway in response to stress stimuli. Biochem. J. 2013;452:531–543. doi: 10.1042/BJ20121886. [DOI] [PubMed] [Google Scholar]
- 23.Bolaños J.P., Almeida A., Moncada S. Glycolysis: a bioenergetic or a survival pathway? Trends Biochem. Sci. 2010;35:145–149. doi: 10.1016/j.tibs.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 24.Cao Y., Zhang X., Wang L., Yang Q., Ma Q., Xu J., et al. PFKFB3-mediated endothelial glycolysis promotes pulmonary hypertension. Proc. Natl. Acad. Sci. U. S. A. 2019;116:13394–13403. doi: 10.1073/pnas.1821401116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rius J., Guma M., Schachtrup C., Akassoglou K., Zinkernagel A.S., Nizet V., et al. NF-kappaB links innate immunity to the hypoxic response through transcriptional regulation of HIF-1alpha. Nature. 2008;453:807–811. doi: 10.1038/nature06905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Obach M., Navarro-Sabaté A., Caro J., Kong X., Duran J., Gómez M., et al. 6-Phosphofructo-2-kinase (pfkfb3) gene promoter contains hypoxia-inducible factor-1 binding sites necessary for transactivation in response to hypoxia. J. Biol. Chem. 2004;279:53562–53570. doi: 10.1074/jbc.M406096200. [DOI] [PubMed] [Google Scholar]
- 27.Borchers A., Pieler T. Programming pluripotent precursor cells derived from Xenopus embryos to generate specific tissues and organs. Genes (Basel) 2010;1:413–426. doi: 10.3390/genes1030413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eto K., Sakura H., Yasuda K., Hayakawa T., Kawasaki E., Moriuchi R., et al. Cloning of a complete protein-coding sequence of human platelet-type phosphofructokinase isozyme from pancreatic islet. Biochem. Biophys. Res. Commun. 1994;198:990–998. doi: 10.1006/bbrc.1994.1141. [DOI] [PubMed] [Google Scholar]
- 29.Rouse J., Cohen P., Trigon S., Morange M., Alonso-Llamazares A., Zamanillo D., et al. A novel kinase cascade triggered by stress and heat shock that stimulates MAPKAP kinase-2 and phosphorylation of the small heat shock proteins. Cell. 1994;78:1027–1037. doi: 10.1016/0092-8674(94)90277-1. [DOI] [PubMed] [Google Scholar]
- 30.Kotlyarov A., Neininger A., Schubert C., Eckert R., Birchmeier C., Volk H.D., et al. MAPKAP kinase 2 is essential for LPS-induced TNF-alpha biosynthesis. Nat. Cell Biol. 1999;1:94–97. doi: 10.1038/10061. [DOI] [PubMed] [Google Scholar]
- 31.Zhao Q., Wang X., Nelin L.D., Yao Y., Matta R., Manson M.E., et al. MAP kinase phosphatase 1 controls innate immune responses and suppresses endotoxic shock. J. Exp. Med. 2006;203:131–140. doi: 10.1084/jem.20051794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Douglas J.J., Walley K.R. Metabolic changes in cardiomyocytes during sepsis. Crit. Care. 2013;17:186. doi: 10.1186/cc13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Waltz P., Carchman E., Gomez H., Zuckerbraun B. Sepsis results in an altered renal metabolic and osmolyte profile. J. Surg. Res. 2016;202:8–12. doi: 10.1016/j.jss.2015.12.011. [DOI] [PubMed] [Google Scholar]
- 34.Iscra F., Gullo A., Biolo G. Bench-to-bedside review: lactate and the lung. Crit. Care. 2002;6:327–329. doi: 10.1186/cc1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Charles C.H., Abler A.S., Lau L.F. cDNA sequence of a growth factor-inducible immediate early gene and characterization of its encoded protein. Oncogene. 1992;7:187–190. [PubMed] [Google Scholar]
- 36.Dean J.L., Sully G., Clark A.R., Saklatvala J. The involvement of AU-rich element-binding proteins in p38 mitogen-activated protein kinase pathway-mediated mRNA stabilisation. Cell Signal. 2004;16:1113–1121. doi: 10.1016/j.cellsig.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 37.Ronkina N., Menon M.B., Schwermann J., Tiedje C., Hitti E., Kotlyarov A., et al. MAPKAP kinases MK2 and MK3 in inflammation: complex regulation of TNF biosynthesis via expression and phosphorylation of tristetraprolin. Biochem. Pharmacol. 2010;80:1915–1920. doi: 10.1016/j.bcp.2010.06.021. [DOI] [PubMed] [Google Scholar]
- 38.Yu H., Sun Y., Haycraft C., Palanisamy V., Kirkwood K.L. MKP-1 regulates cytokine mRNA stability through selectively modulation subcellular translocation of AUF1. Cytokine. 2011;56:245–255. doi: 10.1016/j.cyto.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Manetsch M., Che W., Seidel P., Chen Y., Ammit A.J. MKP-1: a negative feedback effector that represses MAPK-mediated pro-inflammatory signaling pathways and cytokine secretion in human airway smooth muscle cells. Cell Signal. 2012;24:907–913. doi: 10.1016/j.cellsig.2011.12.013. [DOI] [PubMed] [Google Scholar]
- 40.Kirk S.G., Murphy P.R., Wang X., Cash C.J., Barley T.J., Bowman B.A., et al. Knockout of MAPK phosphatase-1 exaggerates type I IFN response during systemic Escherichia coli infection. J. Immunol. 2021;206:2966–2979. doi: 10.4049/jimmunol.2001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hammer M., Mages J., Dietrich H., Servatius A., Howells N., Cato A.C., et al. Dual specificity phosphatase 1 (DUSP1) regulates a subset of LPS-induced genes and protects mice from lethal endotoxin shock. J. Exp. Med. 2006;203:15–20. doi: 10.1084/jem.20051753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chi H., Barry S.P., Roth R.J., Wu J.J., Jones E.A., Bennett A.M., et al. Dynamic regulation of pro- and anti-inflammatory cytokines by MAPK phosphatase 1 (MKP-1) in innate immune responses. Proc. Natl. Acad. Sci. U. S. A. 2006;103:2274–2279. doi: 10.1073/pnas.0510965103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Salojin K.V., Owusu I.B., Millerchip K.A., Potter M., Platt K.A., Oravecz T. Essential role of MAPK phosphatase-1 in the negative control of innate immune responses. J. Immunol. 2006;176:1899–1907. doi: 10.4049/jimmunol.176.3.1899. [DOI] [PubMed] [Google Scholar]
- 44.Abraham S.M., Lawrence T., Kleiman A., Warden P., Medghalchi M., Tuckermann J., et al. Antiinflammatory effects of dexamethasone are partly dependent on induction of dual specificity phosphatase 1. J. Exp. Med. 2006;203:1883–1889. doi: 10.1084/jem.20060336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Burmistrova O., Olias-Arjona A., Lapresa R., Jimenez-Blasco D., Eremeeva T., Shishov D., et al. Targeting PFKFB3 alleviates cerebral ischemia-reperfusion injury in mice. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-48196-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Emini Veseli B., Perrotta P., Van Wielendaele P., Lambeir A.M., Abdali A., Bellosta S., et al. Small molecule 3PO inhibits glycolysis but does not bind to 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase-3 (PFKFB3) FEBS Lett. 2020;594:3067–3075. doi: 10.1002/1873-3468.13878. [DOI] [PubMed] [Google Scholar]
- 47.Liu Y., Shepherd E.G., Nelin L.D. MAPK phosphatases--regulating the immune response. Nat. Rev. Immunol. 2007;7:202–212. doi: 10.1038/nri2035. [DOI] [PubMed] [Google Scholar]
- 48.Frazier W.J., Wang X., Wancket L.M., Li X.A., Meng X., Nelin L.D., et al. Increased inflammation, impaired bacterial clearance, and metabolic disruption after gram-negative sepsis in Mkp-1-deficient mice. J. Immunol. 2009;183:7411–7419. doi: 10.4049/jimmunol.0804343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li J., Wang X., Ackerman W.E.t., Batty A.J., Kirk S.G., White W.M., et al. Dysregulation of lipid metabolism in mkp-1 deficient mice during gram-negative sepsis. Int. J. Mol. Sci. 2018;19:3904. doi: 10.3390/ijms19123904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Heydasch U., Kessler R., Warnke J.P., Eschrich K., Scholz N., Bigl M. Functional diversity of PFKFB3 splice variants in glioblastomas. PLoS One. 2021;16 doi: 10.1371/journal.pone.0241092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Atsumi T., Nishio T., Niwa H., Takeuchi J., Bando H., Shimizu C., et al. Expression of inducible 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase/PFKFB3 isoforms in adipocytes and their potential role in glycolytic regulation. Diabetes. 2005;54:3349–3357. doi: 10.2337/diabetes.54.12.3349. [DOI] [PubMed] [Google Scholar]
- 52.Andersson K., Sundler R. Posttranscriptional regulation of TNFalpha expression via eukaryotic initiation factor 4E (eIF4E) phosphorylation in mouse macrophages. Cytokine. 2006;33:52–57. doi: 10.1016/j.cyto.2005.11.017. [DOI] [PubMed] [Google Scholar]
- 53.Yang K., Fan M., Wang X., Xu J., Wang Y., Tu F., et al. Lactate promotes macrophage HMGB1 lactylation, acetylation, and exosomal release in polymicrobial sepsis. Cell Death Differ. 2022;29:133–146. doi: 10.1038/s41418-021-00841-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zheng Z., Ma H., Zhang X., Tu F., Wang X., Ha T., et al. Enhanced glycolytic metabolism contributes to cardiac dysfunction in polymicrobial sepsis. J. Infect. Dis. 2017;215:1396–1406. doi: 10.1093/infdis/jix138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang T., Lu X., Arnold P., Liu Y., Baliga R., Huang H., et al. Mitogen-activated protein kinase phosphatase-1 inhibits myocardial TNF-alpha expression and improves cardiac function during endotoxemia. Cardiovasc. Res. 2012;93:471–479. doi: 10.1093/cvr/cvr346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li C., Wang Y., Li Y., Yu Q., Jin X., Wang X., et al. HIF1α-dependent glycolysis promotes macrophage functional activities in protecting against bacterial and fungal infection. Sci. Rep. 2018;8:3603. doi: 10.1038/s41598-018-22039-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Colegio O.R., Chu N.Q., Szabo A.L., Chu T., Rhebergen A.M., Jairam V., et al. Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature. 2014;513:559–563. doi: 10.1038/nature13490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kwon S.J., Song J.J., Lee Y.J. Signal pathway of hypoxia-inducible factor-1alpha phosphorylation and its interaction with von Hippel-Lindau tumor suppressor protein during ischemia in MiaPaCa-2 pancreatic cancer cells. Clin. Cancer Res. 2005;11:7607–7613. doi: 10.1158/1078-0432.CCR-05-0981. [DOI] [PubMed] [Google Scholar]
- 59.Yang R., Wilcox D.M., Haasch D.L., Jung P.M., Nguyen P.T., Voorbach M.J., et al. Liver-specific knockdown of JNK1 up-regulates proliferator-activated receptor gamma coactivator 1 beta and increases plasma triglyceride despite reduced glucose and insulin levels in diet-induced obese mice. J. Biol. Chem. 2007;282:22765–22774. doi: 10.1074/jbc.M700790200. [DOI] [PubMed] [Google Scholar]
- 60.Dixit D., Ghildiyal R., Anto N.P., Sen E. Chaetocin-induced ROS-mediated apoptosis involves ATM-YAP1 axis and JNK-dependent inhibition of glucose metabolism. Cell Death Dis. 2014;5:e1212. doi: 10.1038/cddis.2014.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhao Q., Shepherd E.G., Manson M.E., Nelin L.D., Sorokin A., Liu Y. The role of mitogen-activated protein kinase phosphatase-1 in the response of alveolar macrophages to lipopolysaccharide: attenuation of proinflammatory cytokine biosynthesis via feedback control of p38. J. Biol. Chem. 2005;280:8101–8108. doi: 10.1074/jbc.M411760200. [DOI] [PubMed] [Google Scholar]
- 62.Dorfman K., Carrasco D., Gruda M., Ryan C., Lira S.A., Bravo R. Disruption of the erp/mkp-1 gene does not affect mouse development: normal MAP kinase activity in ERP/MKP-1-deficient fibroblasts. Oncogene. 1996;13:925–931. [PubMed] [Google Scholar]
- 63.Lee J.C., Laydon J.T., McDonnell P.C., Gallagher T.F., Kumar S., Green D., et al. A protein kinase involved in the regulation of inflammatory cytokine biosynthesis. Nature. 1994;372:739–746. doi: 10.1038/372739a0. [DOI] [PubMed] [Google Scholar]
- 64.Zhang T., Inesta-Vaquera F., Niepel M., Zhang J., Ficarro S.B., Machleidt T., et al. Discovery of potent and selective covalent inhibitors of JNK. Chem. Biol. 2012;19:140–154. doi: 10.1016/j.chembiol.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim V.Y., Batty A., Li J., Kirk S.G., Crowell S.A., Jin Y., et al. Glutathione reductase promotes fungal clearance and suppresses inflammation during systemic Candida albicans infection in mice. J. Immunol. 2019;203:2239–2251. doi: 10.4049/jimmunol.1701686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shepherd E.G., Zhao Q., Welty S.E., Hansen T.N., Smith C.V., Liu Y. The function of mitogen-activated protein kinase phosphatase-1 in peptidoglycan-stimulated macrophages. J. Biol. Chem. 2004;279:54023–54031. doi: 10.1074/jbc.M408444200. [DOI] [PubMed] [Google Scholar]
- 67.Wang X.X., Zhao Q., Matta R., Meng X.M., Liu X.P., Liu C.G., et al. Inducible nitric-oxide synthase expression is regulated by mitogen-activated protein kinase phosphatase-1. J. Biol. Chem. 2009;284:27123–27134. doi: 10.1074/jbc.M109.051235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee S.Y., Buhimschi I.A., Dulay A.T., Ali U.A., Zhao G., Abdel-Razeq S.S., et al. IL-6 trans-signaling system in intra-amniotic inflammation, preterm birth, and preterm premature rupture of the membranes. J. Immunol. 2011;186:3226–3236. doi: 10.4049/jimmunol.1003587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Numata M., Morinaga S., Watanabe T., Tamagawa H., Yamamoto N., Shiozawa M., et al. The clinical significance of SWI/SNF complex in pancreatic cancer. Int. J. Oncol. 2013;42:403–410. doi: 10.3892/ijo.2012.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data have been deposited in the Gene Expression Omnibus (GSE122741) https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE122741.