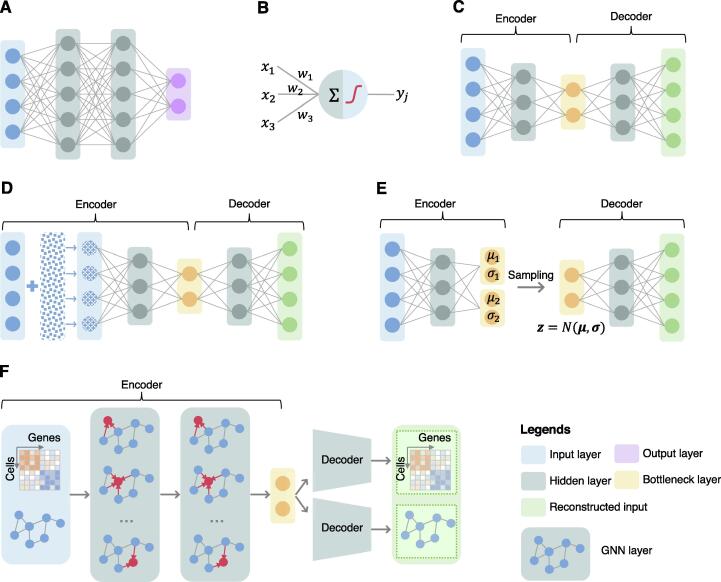
Figure 2.
Illustration of deep learning architectures that have been used in scRNA-seq analysis
A. Basic design of a feed-forward neural network. B. A neural network is composed of “neurons” organized into layers. Each neuron combines a set of weights from the prior layer, and passes the weighted summed value through a non-linear activation function, such as sigmoid, rectifier (i.e., ReLU), and hyperbolic tangent, to produce a transformed output. C. Autoencoder, a special variant of the feed-forward neural network aiming at learning low-dimensional representations of data while preserving data information. D. DAE, a variant of autoencoder, which was developed to address overfitting problems of autoencoders. DAE forces the input data to be partially corrupted and tries to reconstruct the raw un-corrupted data. E. VAE, a variant of autoencoder, aiming at compressing input data into a constrained multivariate latent distribution space in the encoder, which is regular enough and can be used to generate new content in the decoder. F. GAE. Benefiting from the advanced deep learning architecture GNN, GAE has been developed and used in scRNA-seq analysis. The encoder of GAE considers both sample features (e.g., the gene expression profiles/counts of cells) and samples’ neighborhood information (e.g., topological structure of cellular interaction network) to produce low-dimensional representations while preserving topology in data. The decoder unpacks the low-dimensional representations to reconstruct the input network structure and/or sample features. ReLU, rectified linear unit; DAE, denoising autoencoder; VAE, variational autoencoder; GAE, graph autoencoder; GNN, graph neural network.