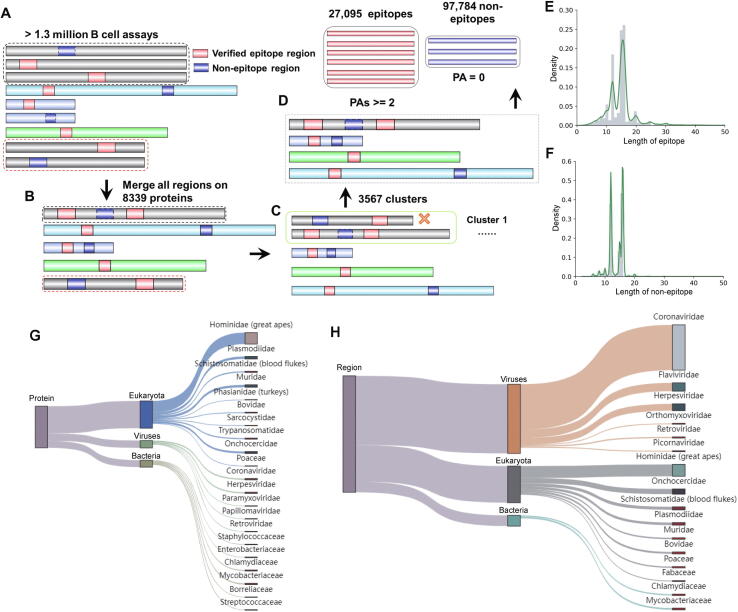
Figure 1.
Benchmark data preparation and evaluation
A. The experimentally identified epitope-containing regions were collected from the IEDB database. B. Identical protein sequences were integrated and the verified epitope regions were aggregated. C. Sequence redundancy was cleaned for the similar proteins by CD-HIT. D. Proteins with the largest number of epitope-containing regions were retained. The curated dataset was divided into epitopes and non-epitopes according to epitope assay information. We defined all epitope-containing regions that were tested by at least two PAs as epitopes to avoid possible chance of a single test result. Moreover, all epitope-containing regions that were tested in at least two assays but not tested as positive in any assay were stored as non-epitopes. All other epitope-containing regions with inconsistent test responses that did not meet both criteria were excluded. E. The length distribution of epitopes. F. The length distribution of non-epitopes. G. Taxonomic distribution in super-kingdoms and families at the protein level. H. Taxonomic distribution in super-kingdoms and families at the verified epitope level. PA, positive assay.