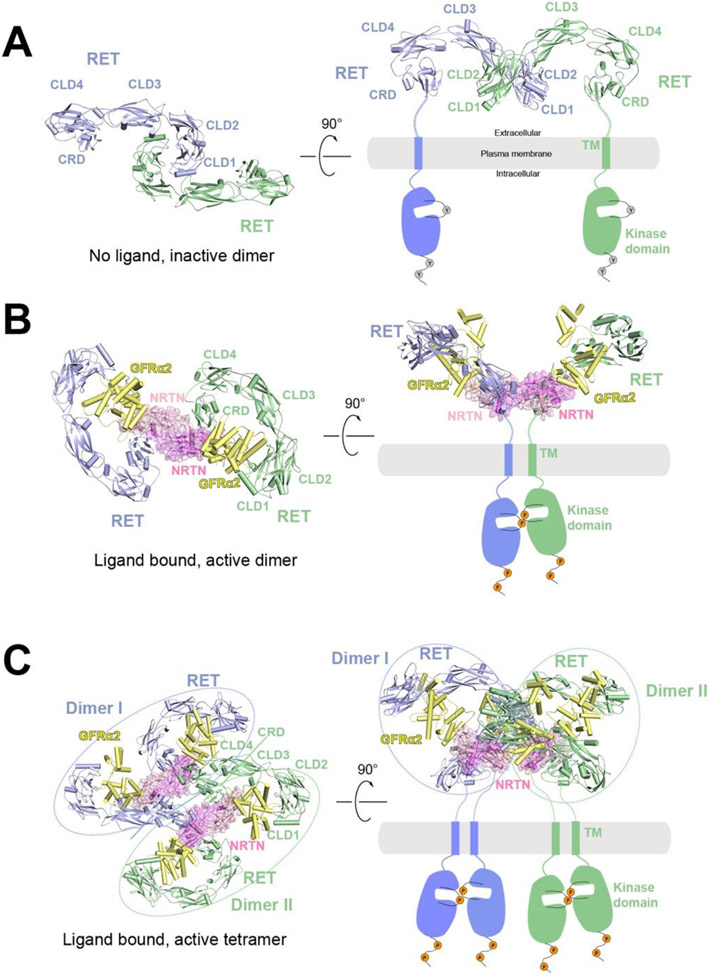
Figure 2.
Overall structures of RET at different oligomerization states and proposed activation mechanisms. The protomers in the receptor dimer or tetramer (blue and green) as well as the co-receptor (yellow) are shown in cartoon representations, whereas the ligands are shown in cartoon and surface (pink) representations with semi-transparency. The TM and kinase domains, not present in the structure, are indicated by schematic drawings.
A) Structural model of RET in apo state as an inactive dimer in two different views. The structural model is generated based on the crystal structure of CLD1/CLD2 domains (PDB: 2X2U). In this structural model, the two CRD domains are far apart, suggesting an autoinhibited state.
B) Cryo-EM structure of the 2:2:2 NRTN/GFRα2/RET complex in two different views (PDB: 6Q2O). In this structure, the two CRD domains are close to each other, consistent with being an active dimer.
C) Cryo-EM structure of the 4:4:4 NRTN/GFRα2/RET complex in two different views (PDB: 6Q2R). Dimer I (blue) and Dimer II (green) are structurally identical to the 2:2:2 NRTN/GFRα2/RET complex.