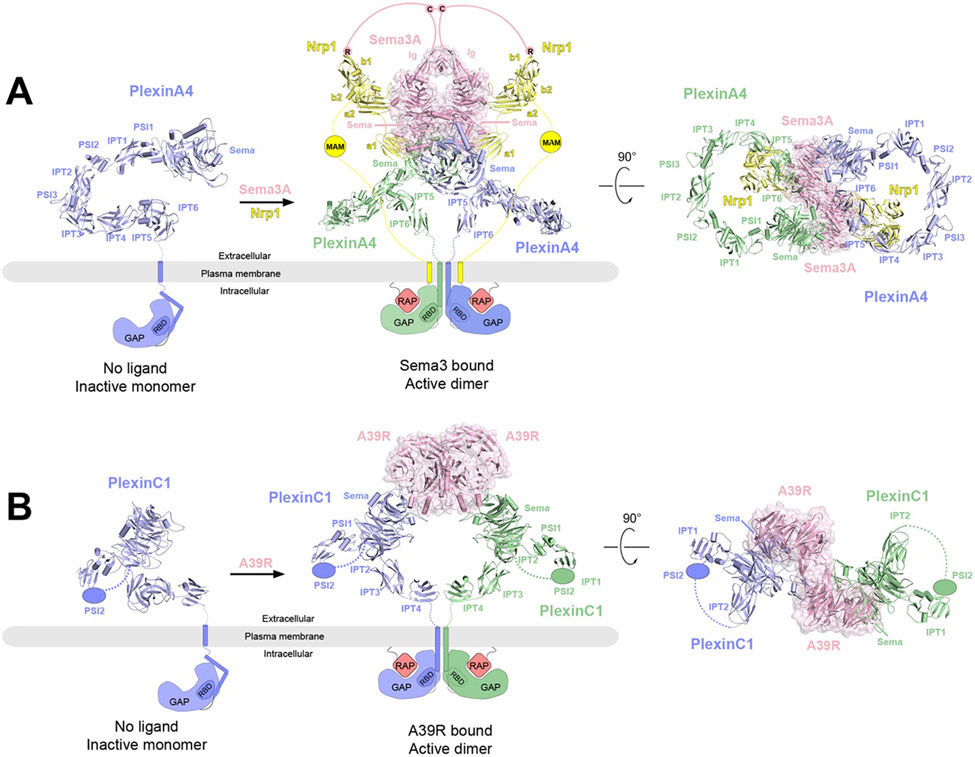
Figure 5.
Overall structures of PlexinA4 and PlexinC1 in their respective apo and ligand-bound forms and the proposed activation mechanisms. The protomers in the receptor dimers (blue and green) as well as the coreceptor Nrp1 (yellow) are shown in cartoon representations, whereas the ligands are shown in cartoon and surface (pink) representations with semi-transparency. The missing domains and linkers in each structure are shown as schematic representations. In the apo state, the GAP domains of PlexinA4 or PlexinC1 are in the autoinhibited state with the Rap-binding site inaccessible. Binding of the ligands (Sema3A or A39R) induces plexin dimerization, opening up the GAP active site to allow Rap binding and GTP hydrolysis.
A) Left, crystal structure of ECD of PlexinA4 in its apo state (PDB: 5L5K). Middle and right, cryo-EM structure of 2:2:2 PlexinA4/Nrp1/Sema3A complex in two different views (PDB: 7M0R). The structure of the ECD of PlexinA4 in both structures are essentially the same. The lines represent the various linker domains between different protein components that are important to the formation of the complex. The pink lines represent the linker between C723 and the Ig-like domains of Sema3A as well as that between C723 and the C-terminal R770, which engages the binding pocket in the b1 domain of Nrp1. The yellow lines represent the b2-MAM and MAM-TM linkers in Nrp1.
B) Left, structural model of monomeric PlexinC1 in its apo state taken from the structure of the PlexinC1/A39R complex (PDB: 6VXK). Middle and right, the cryo-EM structure of the 2:2 PlexinC1/A39R complex (PDB: 6VXK) in two different views.