Figure 1.
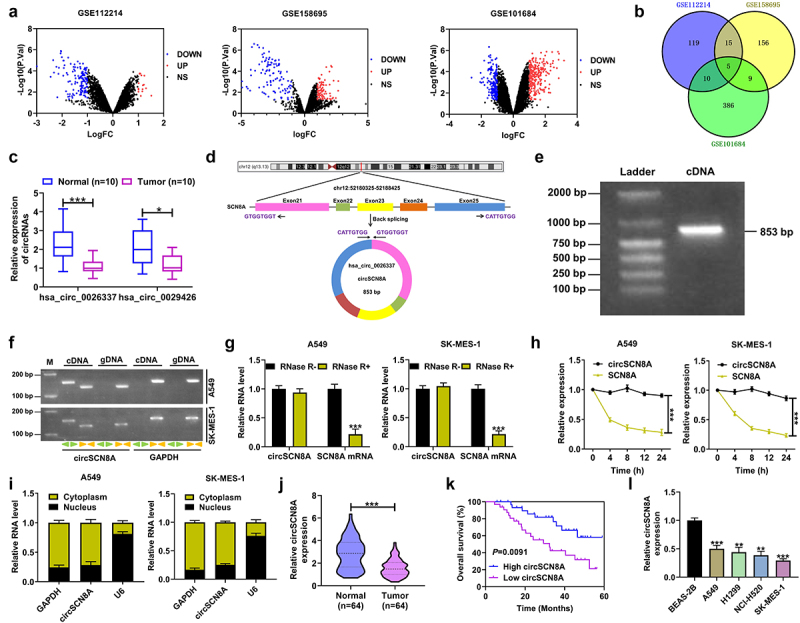
Expression and characterization of circSCN8A in NSCLC. (a) Volcano map showing the DECs in NSCLC according to the microarray data (GSE112214, GSE158695, and GSE101684). (b) Venn diagram showing five commonly down-regulated circRnas in GSE112214, GSE158695, and GSE101684. (c) the expression of hsa_circ_0026337 and hsa_circ_0029426 was determined by qRT-PCR in 10 pairs of NSCLC tissues and adjacent normal tissues. (d) Schematic diagram showing the circularization of hsa_circ_0026337 by back-splicing of SCN8A exons 21–25. (e) Detection of circSCN8A by agarose gel electrophoresis. (f) PCR was performed to amplify circSCN8A and its linear counterpart SCN8A by using convergent or divergent primers with cDNA or gDNA in NSCLC cells as the template. GAPDH acted as a control. (g and h) qRT-PCR analysis was used to assess the expression of circSCN8A and SCN8A mRNA in A549 and SK-MES-1 cells after treatment with RNase and actinomycin D. (i) Subcellular localization of circSCN8A in A549 and SK-MES-1 cells. (j) CircSCN8A expression in NSCLC tissues (n = 64) and corresponding paracancer tissues (n = 64). (k) Kaplan-Meier survival curve was plotted to evaluate the correlation between circSCN8A expression and overall survival of NSCLC patients. (l) CircSCN8A expression in NSCLC cells. *P <0.05, **P <0.01, ***P <0.001.
