Figure 5. Versatility and performance of BAMM Prep method.
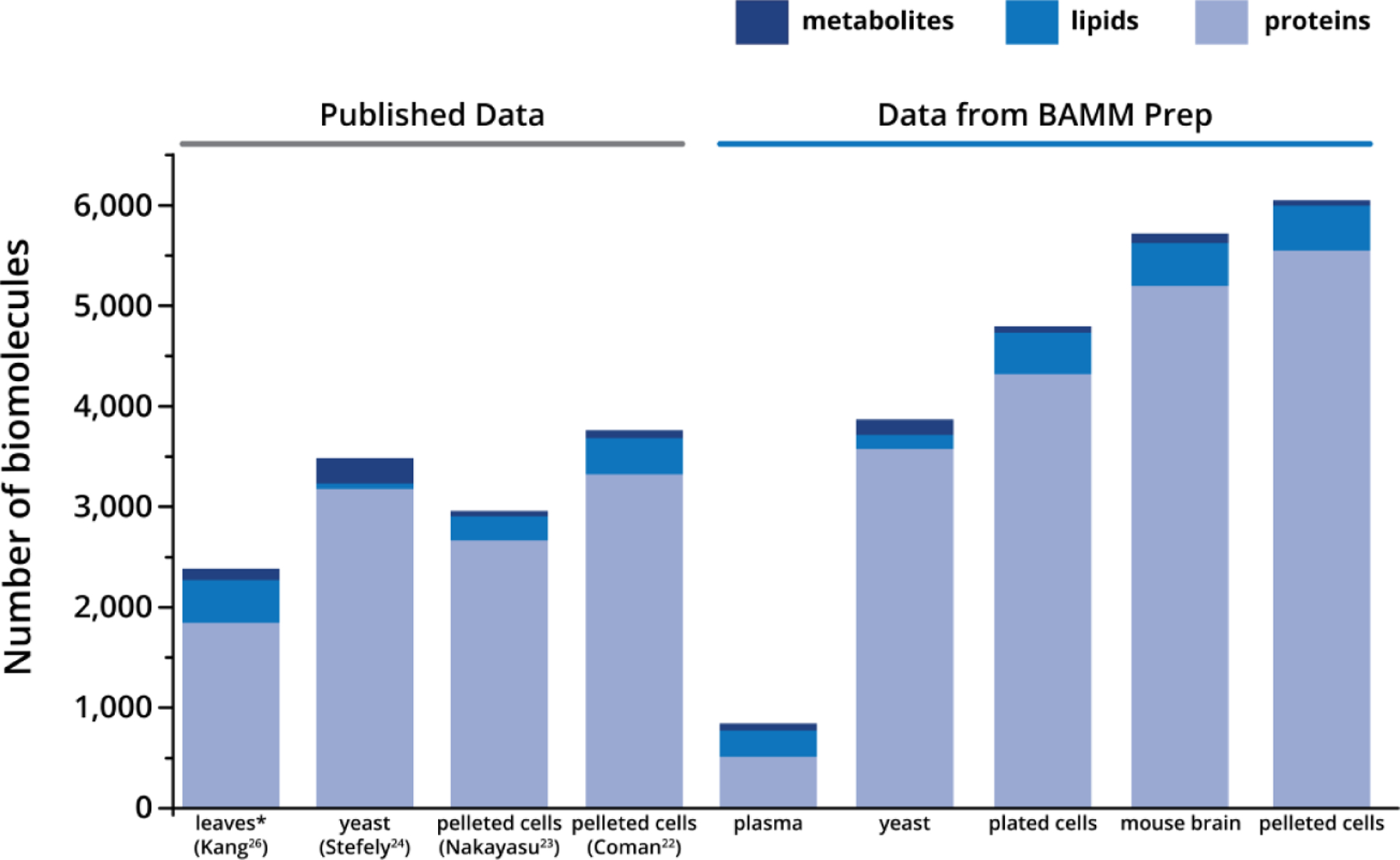
The four lefthand columns (“Published Data”) show the number of proteins, lipids, and metabolites identified from published multi-omic sample preparation workflows. All numbers were taken as reported in the results sections of the publications; however, the reported number of metabolites in Kang et al. was reduced to only those that matched the criteria for metabolite identifications used in this manuscript. The five righthand columns (“Data from BAMM Prep”) show the number of identified proteins, lipids, and metabolites from our streamlined multi-omic sample preparation (numbers are the average of three replicates). The specific samples are as follows: A. thaliana (Kang), S. cerevisiae (Stefely), Calu-3 cells (Nakayasu), mesenchymal stem cells (Coman), and NIST 1950 plasma, S. cerevisiae, cultured mouse adipocytes, C57BL/6J mouse brain, HEK293 (BAMM Prep)
