Figure 3. Cas8c N-term has a minimal PAM-recognition requirement.
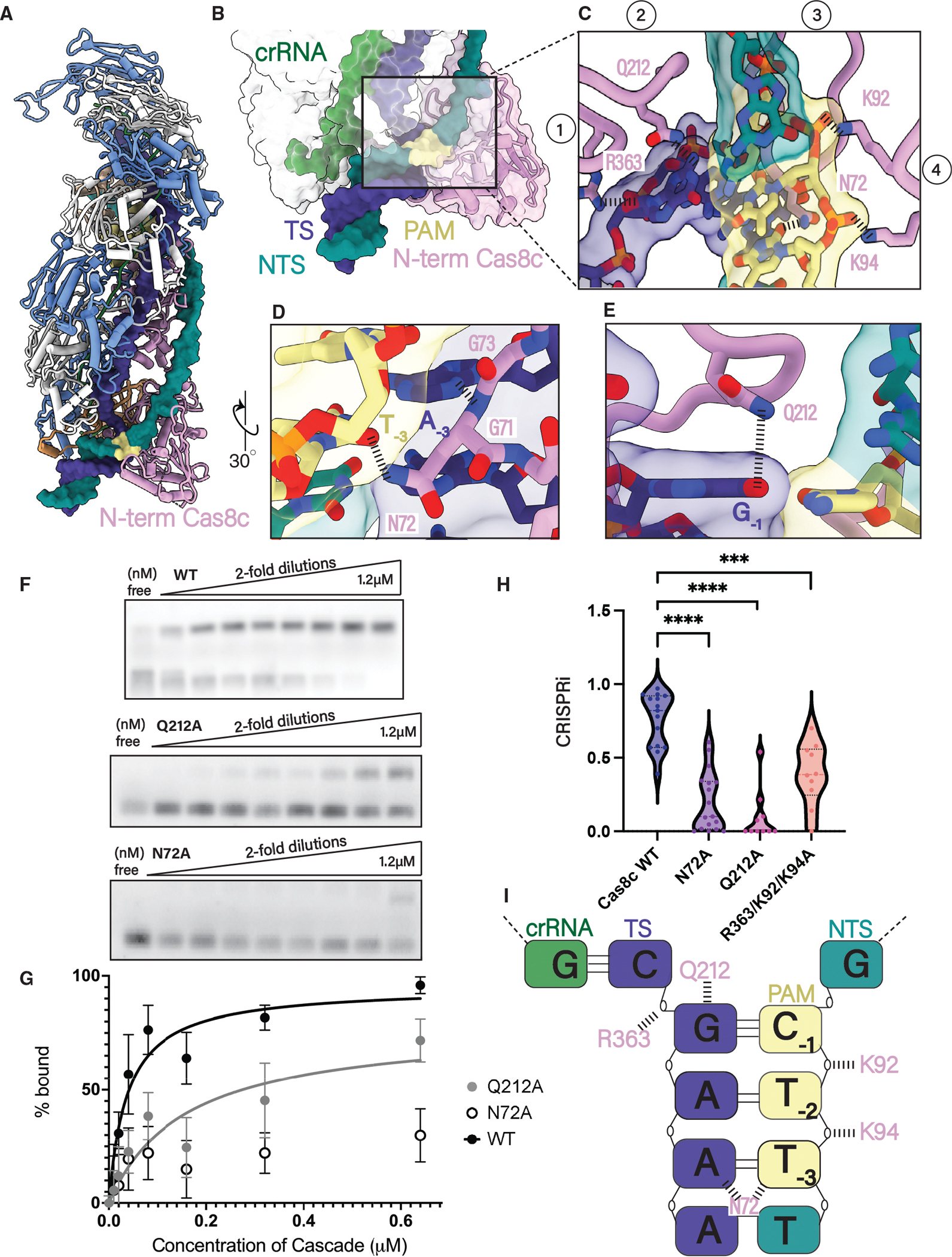
(A) 5′-TTC PAM motif is recognized from the minor grove by the N-term of the Cas8c large subunit.
(B and C) (B) The N-term of Cas8c clamps around the dsDNA helix and (C) positions four loops to facilitate PAM recognition and strand separation.
(D) N72 makes one contact with the PAM, protruding from a glycine loop that wedges into the duplex.
(E) dsDNA splitting is facilitated by a glutamine wedge that stacks above the PAM and intercalates between the two DNA strands.
(F) Electrophoretic mobility shift assays (EMSAs) measuring the binding of increasing concentrations of Cascade to a fluorescently labeled dsDNA target with a 5′-TTC PAM. Experiments were done in triplicate and representative results are shown.
(G) Quantification of EMSA data. Each point is the average of at least three independent replicates. WT Cascade Kd(app) is 37 nM, and Q212A mutant Kd(app) is 155 μM. The binding curve of the N72A mutant could not be fit and a Kd could not be determined.
(H) Quantification of type I-C Cascade in vivo plasmid interference assay with PAM mutants. Each data point is an independent replicate. PAM mutants Q212A and N72A demonstrated the most severe defect in interference efficiency compared with WT and the R363A/K92A/K94A mutant. **** demonstrates mutants are statistically significant to WT (N72A: p < 0.0001, Q212A: p < 0.0001, R363A/K92A/K94A: p = 0.0003).
(I) Schematic of the five residues involved in PAM recognition for type I-C Cascade. Two residues (Q212 and N72) make specific contact with the PAM bases and three residues (K92, K94, and R363) stabilize the phosphate backbone.
