Figure 4. AcrIF2 blocks both PAM recognition and domain rearrangements required for R-loop formation.
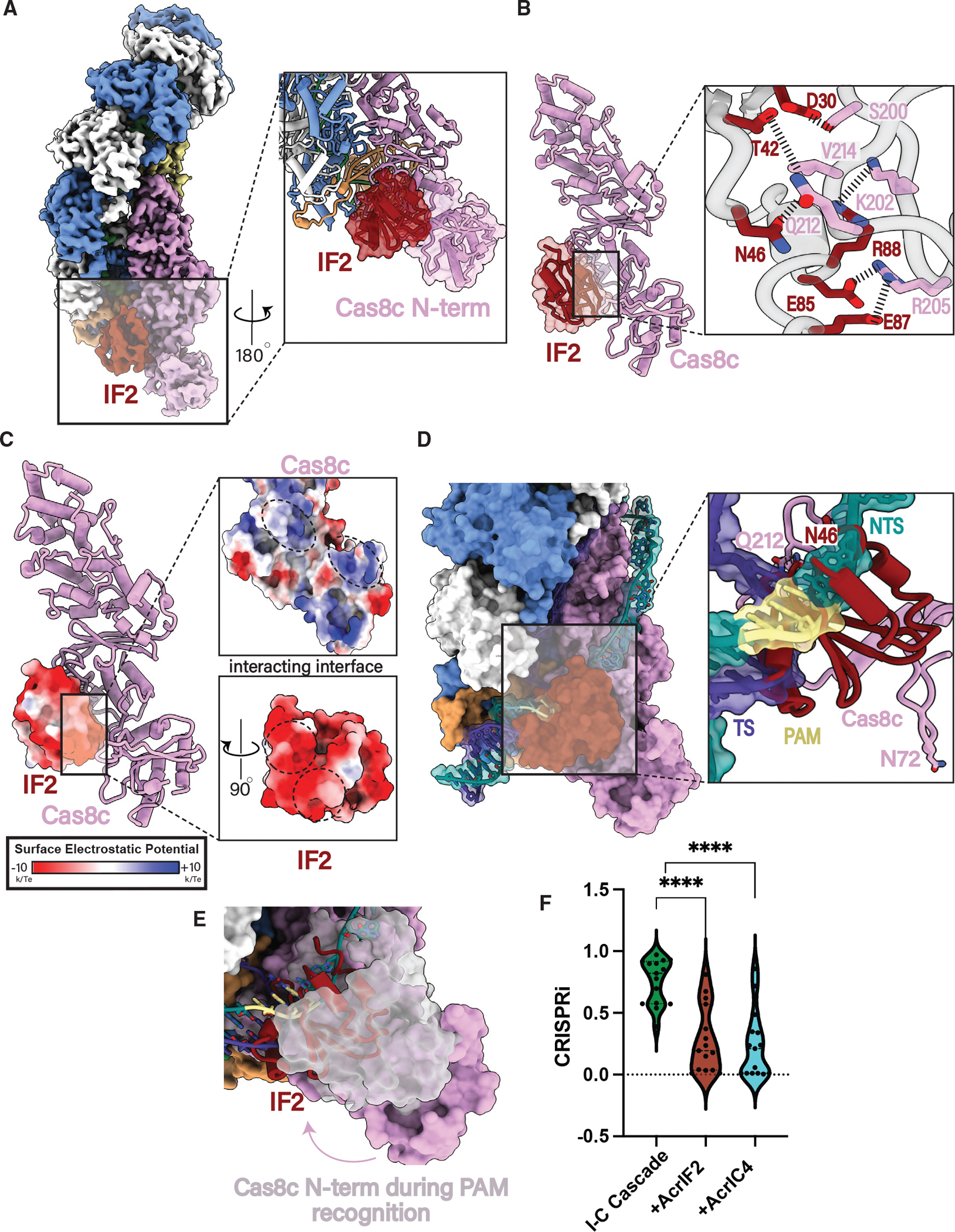
(A) 3.0 Å resolution cryoelectron structure of type I-C Cascade bound to AcrIF2 and atomic model demonstrating the presence of Cas8c N-term (inset).
(B) AcrIF2 exclusively interacts with Cas8c through a large network of non-specific interactions.
(C) The surface of AcrIF2 is negatively charged and sits within two positively charged surfaces between the Cas8c N-term and the rest of the Cas8c subunit.
(D) Structural superposition of the type I-C dsDNA-bound model shows severe clashing between PAM residues and AcrIF2. AcrIF2 additionally hinders the rearrangement of Cas8c N-term containing important residues involved in PAM recognition (N72).
(E) AcrIF2 occludes PAM recognition by Cas8c N-terminal domain. Pink, AcrIF2-bound structure; gray, full R-loop structure.
(F) Type I-C Cascade-Cas3 in vivo plasmid interference assays demonstrate AcrsIF2 and AcrIC4 inhibit DNA degradation. **** demonstrate AcrIF2 and AcrIC4 are statistically significant (AcrIF2: p < 0.0001, AcrC4: p < 0.0001).
