Abstract
Background:
Vulvodynia (idiopathic vulvar pain) affects up to 8% of women by age 40, has a poorly understood etiology, and variable treatment efficacy. Several risk factors are associated with vulvodynia from history of yeast infections to depression and allergies. Recent work suggests an altered immune inflammatory mechanism plays a role in vulvodynia pathophysiology. As the vaginal microbiome plays an important role in local immune-inflammatory responses, we evaluated the vaginal microbiome among women with vulvodynia compared to controls as one component of the immune system.
Objective:
Characterize the vaginal microbiome in women with clinically-confirmed vulvodynia and age-matched controls and assess its overall association with vulvodynia, and how it may serve to modify other factors that are associated with vulvodynia as well.
Study Design:
We conducted a case-control study of 234 Minneapolis/Saint Paul area women with clinically-confirmed vulvodynia and 234 age-matched controls clinically confirmed with no history of vulvar pain. All participants provided vulvovaginal swab samples for culture-based and non-culture (sequencing) based microbiological assessments; background and medical history questionnaires on demographic characteristics, sexual and reproductive history, and history of psychosocial factors. Vaginal microbiome diversity was assessed using the Shannon Alpha Diversity Index. Data were analyzed using logistic regression.
Results:
Culture and molecular-based analyses of the vaginal microbiome showed few differences between cases and controls. However, among women with alpha diversity below the median (low), there was a strong association between increasing numbers of yeast infections and vulvodynia onset, relative to comparable time periods among controls (age-adjusted odds ratio [OR] 8.1, 95% CI: 2.9–22.7 in those with 5 or more yeast infections). Also among women with low diversity microbiomes, we observed a strong association between moderate-to-severe childhood abuse, antecedent anxiety, depression, and high levels of rumination and vulvodynia with ORs from 1.83 to 2.81. These associations were not observed in women with high diversity microbiomes.
Conclusions:
Although there were no overall differences in microbiome profiles between cases and controls, vaginal microbiome diversity influenced associations between environmental and psychosocial risk factors and vulvodynia. However, it is unclear whether vaginal diversity modifies the association between the risk factors and vulvodynia or is altered as a consequence of the associations.
Keywords: case-control studies, vulvodynia, yeast infections, microbiome
Condensation:
The associations between history of yeast infections, anxiety, and childhood abuse and vulvodynia may be modified by characteristics of the vaginal microbiome.
Introduction
Vulvodynia is defined as idiopathic vulvar pain of at least 3 months’ duration and is estimated to affect up to 8% of women during their reproductive years (1,2). Vulvodynia is distinct from vulvar pain related to inflammation, neoplasm, or traumatic injury (3). Proposed risk factors can be grouped broadly into pathophysiologic (e.g. hormonal, inflammatory, environmental) and psychosocial factors (e.g. mood, childhood victimization, and other psychosocial events) and etiology likely involves a combination of biopsychosocial exposures that impact a heterogeneous patient population (3).
Previous research suggests an altered immune-inflammatory response may be an important pathogenic mechanism (4). Both clinical- and laboratory-based studies have found associations between vulvodynia and increased production of the pro-inflammatory cytokine interleukin-1β (5, 6). Further research by Harlow et al. (7) found that women with vulvodynia were 2.5 times more likely to self-report hives prior to first report of vulvar pain relative to a comparable exposure time period among controls (95% confidence interval [CI], 1.7–4.4). Further, women with vulvodynia were 5.5 (1.7–17.8) times more likely to report 10 or more yeast infections during their lifetime before vulvodynia onset relative to controls during a comparable time period after adjusting for age, age at first sexual intercourse and antecedent urinary tract infections (8).
The vaginal microbiome plays an important role in local immune-inflammatory responses (9). A more diverse vaginal microbiome is associated with increased inflammatory cytokine levels and Lactobacillus-dominated microbiomes with decreased proinflammatory cytokine levels (10). A recent study of 30 women with vulvar vestibulitis syndrome (VVS) and 15 controls showed no statistically significant vaginal microbiome differences (11), but there were varying rates of colonization with candida, lactobacillus species, and streptococcus. A second study assessed only culture-based differences between 50 cases of vulvodynia and 50 clinic-based controls and reported a reduction in the diversity of Lactobacilli and greater prevalence of candida and other fungi among cases compared to controls (12). Our substantially larger study expands on previous work, using a community-based sample of clinically confirmed cases of vulvodynia and controls matched on cases’ age at diagnosis of vulvodynia.
MATERIALS AND METHODS
Study design/study population
We analyzed data and samples from a case-control study of vulvodynia among women 18–40 years old described previously (1,8). Briefly, ~30,000 women completing a screener survey at one of approximately 40 community health clinics in the Minneapolis/Saint Paul metropolitan area between March 2010 and October 2013, were eligible. Those likely to meet the International Society for the Study of Vulvovaginal Diseases (ISSVD) criteria for vulvodynia and randomly selected controls with no history of vulvar discomfort were invited to participate in a clinical visit. Of the approximately 1,400 women invited for further vulvar pain assessment, 350 completed their examination and 234 were clinically confirmed as cases of vulvodynia. Of the 2,287 control women invited, 251 agreed to participate and 234 were clinically confirmed as having no ongoing or past history of vulvar pain. Controls who were the same age or older as a case’s age at first onset of vulvar pain were randomly matched to cases and assigned a “reference age” identical to the age at first onset of vulvar pain in the matched case. For example, a 30 year old woman with vulvodynia onset at age 25 was matched to a woman with no history of vulvodynia who was at least 25 years of age. “25” was then considered the “reference age” and certain exposures of interest were obtained prior to that reference age for both cases and controls.
Data Collection: Outcome and Exposure Assessments
All participants completed the clinical visit which followed a standardized approach to confirm the presence or absence of vulvodynia, including a careful medical history, a physical examination with pH assessment, wet prep, and cultures to rule out vaginitis, especially candidiasis, dermatosis, irritants or allergens. In keeping with the diagnostic criteria set by the ISSVD and the most recent NIH consensus conference on vulvodynia, the diagnosis of vulvodynia was made by the following: a history of vulvovaginal pain, spontaneous or elicited by touch, and skin findings that are limited to erythema of the vulva or vagina (13). Participants could not have an active yeast infection defined by symptoms of burning, pruritis or vulvar irritation accompanied by documented yeast in the vaginal fluid by microscopy or culture at the time of study enrollment. Cases and controls completed several structured questionnaires to provide detailed information about their reproductive and medical history, personal hygiene, and psychosocial factors that preceded the onset of their vulvar pain or comparable age period among controls.
Microbial Collection and Laboratory Methods
A vulvar specialist used three Dacron cotton swabs to collect vaginal fluid samples by inserting the swabs simultaneously into the introitus about 1 inch, and then rotating 5 times. The first two swabs were used for quantitative vaginal cultures and were inserted into a Port-A-Cul transport tube, then capped and shipped to the SH Laboratory at Magee-Womens Research Institute. The third swab was inserted into a cryovial, labeled and then stored at −70 degrees centigrade. No media or buffer was used in this tube. This specimen was shipped to the BF laboratory and used for 16S rRNA sequencing. A detailed description of the methods for obtaining microbial counts is described in the supplementary methods (Appendix S1). In brief, classical culture-based methods were used to quantitate the number of bacteria in the vaginal swab using selective media and standard counting and classification methods (eg Nugent score).
16S rRNA Sequencing and bioinformatics:
DNA extraction was performed using Mag Attract Power Microbiome kit (Qiagen, cat # 27500–4-EP) with glass bead plates. The 16S rRNA V4 region was amplified and sequenced using the Dual indexing sequencing protocol (14). Analysis of the 16S rRNA V4 hypervariable region began with a modified MOTHUR SOP, originally developed by Kozich et al (14; accessed March 2018). During this process, sequences were paired and aligned to the SILVA v.123 reference database. Sequencing was performed on the Illumina MiSeq platform using Miseq Reagent Kit V2 at 500 cycles (Illumina, cat# MS102–2003). In lieu of pre-clustering and clustering via the Mothur SOP, oligotyping was conducted utilizing an unsupervised minimum entropy decomposition (MED) method (15,16). Species level classification was assigned using the open source dada2 v.1.6.0 R package (17). For oligotypes that remained unclassified, representative sequences that uniquely matched an NCBI BLAST+ species level taxonomic suggestion at greater than or equal to 98% identity were assigned. If taxa could not be assigned the oligotype is noted by genus and oligotype number. There were 13,732,126 reads used in the analysis. The range per person was 1,520 to 87,493, with a median of 24,613. In order to reduce the complexity of the count data, we calculated alpha diversity using the Shannon diversity index and used the median alpha diversity to classify cases and controls into high (≥median) and low (<median) diversity groups. Shannon diversity takes into account the relative abundance (that is, the evenness of taxa in the community) and the number of taxa (richness). To assess whether associations with alpha diversity were attributable to differences in evenness and/or richness we categorized cases and controls into high (≥median) and low (<median) groups by evenness and richness.
To identify community state types (CSTs), we used an unsupervised clustering method appropriate for compositional data, Dirichlet multinomial mixture models (18–20); R v.3.4.0 with Dirichlet Multinomial v.1.20.0 package). The number of CSTs was determined by comparing the Laplace approximation of the negative log models and identifying the point at which an increase in Dirichlet components resulted in minor reductions of model fit. As shown in the waffle plot (Figure 2), each square represents a 1% contribution to the average relative abundance across samples assigned to that community state type. Any species that has an average relative abundance of less than 2% in all 6 community state types is grouped into “other” for this figure. We have also provided a heat map (Figure S2) where the colors represent square-root transformed counts, with darker colors representing larger counts. The thicker bands (big blocks of color) are averages across all the samples in that community state type, and these can be used to determine the dominant species in that CST. Further details can be found in Appendix S1.
Figure 2 –
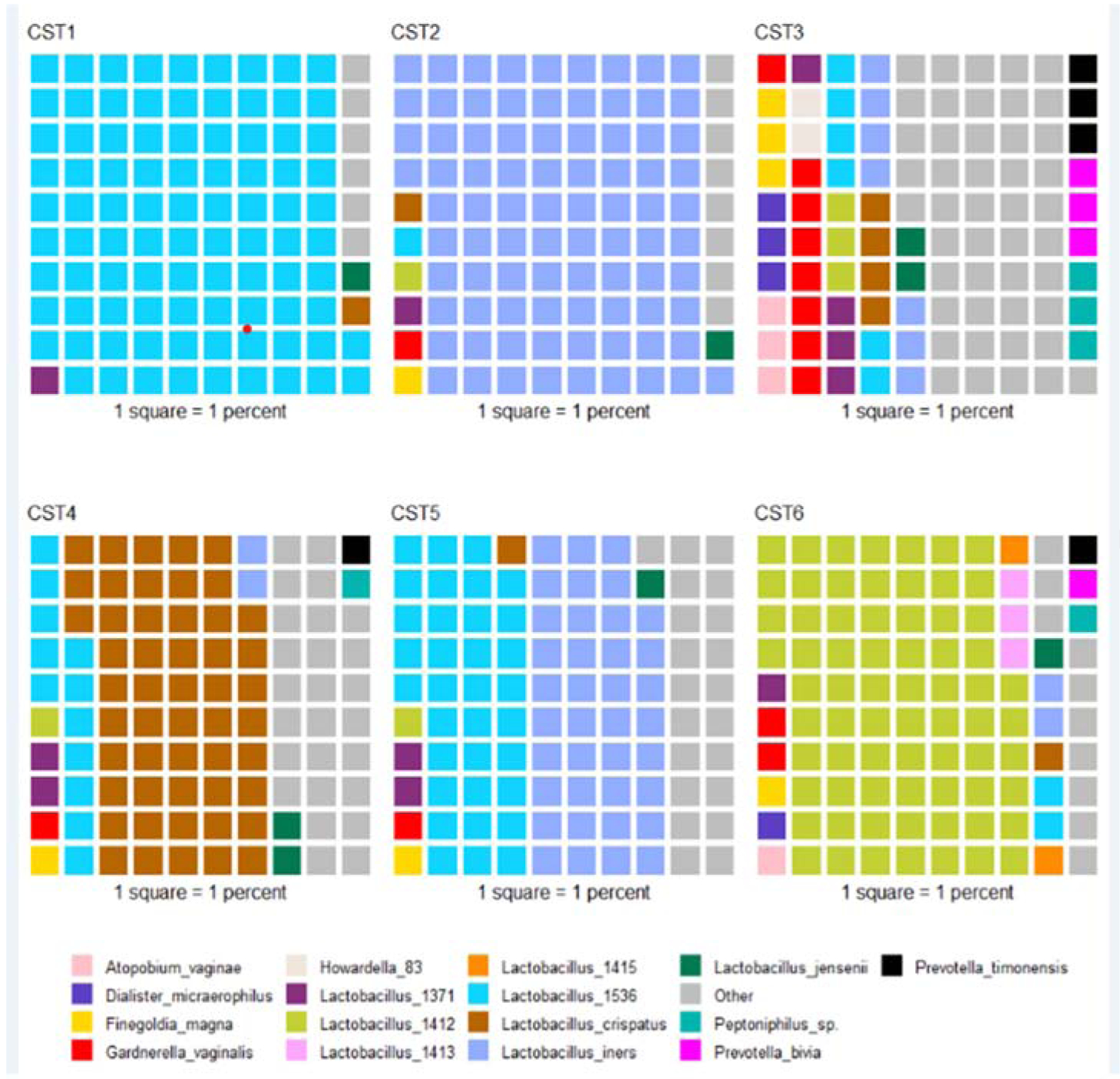
Composition of community state types resulting from clustering of microbiomes across participants using Dirichlet multinomial mixed models. Each square represents 1 percent of the average relative abundance of the samples assigned to that community state type. Taxa whose average relative abundance across all communities state types was less than 2 percent are grouped into ‘other.’ Lactobacillus taxa identified as different oligotypes that could not be assigned to a specific species are noted by number (e.g., Lactobacillus 1536 shown in turquoise).
Lastly, we used ALDEx2 v.1.20.0 (16, 17) to investigate associations of individual taxa abundance by vulvodynia status and community state type. This package transforms taxon abundances for each sample to centered-log ratios and calculates a median centered log ratio for each group of interest. The effect sizes for each taxon calculated by ALDEx2 summarize the ratio of between group differences to within group differences. Statistical significance for effect sizes were calculated using the Wilcoxon rank sum test with false discovery rate corrected p-values using the Benjamini-Hochberg correction. The parameters used for this step included 1000 Monte-Carlo simulations.
Statistical Analyses
Complete microbiome sequencing data were available for 215 cases and 222 controls. For this analysis, the reference age matching was not retained, but assessed as a covariate, particularly because we assessed the impact of the vaginal microbiome as an effect measure modifier (21). We first described demographic, medical, and psychosocial characteristics between those with and without vulvodynia, and the distribution of factors associated with microbial features (for example antibiotic use, autoimmune conditions, history of bacterial vaginosis). We then fit logistic regression models to estimate the odds ratios (OR) and 95% confidence intervals (CI) for selected risk factors and vulvodynia adjusted for age and stratified by Shannon Alpha Diversity above the median (high) and below the median (low). Similar models were fit stratified by categorical community state types (CSTs). CSTs were determined using Dirichlet multinominal mixed models, and unsupervised clustering method described in the Appendix S1. CST1, CST2, and CST3 were treated as separate groups and CST4–6 were combined due to small numbers and similar patterns of diversity (e.g. multiple organism dominant). Analyses were conducted using SAS v9.4.
Funding:
This research was supported by the National Vulvodynia Association and NIH-NICHD R01 HD058608.
RESULTS
Vulvodynia cases were, on average, two years younger than controls but were similar in body mass index (Table 1). Cases reported fewer sexual partners, pregnancies, and years of contraceptive use than controls. As previously shown (8), cases were more likely than controls to report a history of gynecological infections, allergenic exposures (e.g. seasonal allergies, hives, and insect bite sensitivities), autoimmune diseases, and history of other comorbid functional somatic syndromes. Cases were also more likely than controls to report a history of psychosocial and psychological morbidity including a history of childhood abuse, anxiety, depression, and rumination preceding their first onset of vulvar pain.
Table 1:
Demographic characteristics of 215 women with chronic vulvar pain (cases) and 222 women with no vulvar pain history (controls); 2010–2015.
| Cases | Controls | ||
|---|---|---|---|
| N (%) | N (%) | ||
| 215 (49.2) | 222 (50.8) | ||
| White Race | 200 (93.02) | 206 (92.79) | |
| Age (years) | 18–25 | 53 (24.6) | 29 (13.1) |
| 26–30 | 78 (36.2) | 65 (29.3) | |
| 31–35 | 61 (28.3) | 66 (29.7) | |
| 36–40 | 23 (10.7) | 62 (27.9) | |
| BMI | <20 | 28 (13.0) | 20 (9.0) |
| 20–25.9 | 104 (48.3) | 110 (49.6) | |
| 26–30.9 | 48 (22.3) | 44 (19.8) | |
| ≥31 | 35 (16.2) | 48 (21.6) | |
| Sexual and Reproductive History | |||
| Sexual Partners (lifetime) | <5 | 99 (46.0) | 96 (43.2) |
| 5–10 | 58 (26.9) | 46 (20.7) | |
| ≥10 | 58 (26.9) | 80 (36.0) | |
| Years on Hormonal Contraceptive | <5 | 78 (36.2) | 70 (31.5) |
| 5–10 | 67 (31.1) | 61 (27.5) | |
| ≥10 | 70 (32.5) | 91 (41.0) | |
| Ever Pregnant | 103 (47.9) | 130 (58.6) | |
| Nulliparous | 129 (60) | 107 (48.2) | |
| Gynecological Infections 1 | 0 | 115 (53.4) | 145 (65.3) |
| 1 | 66 (30.7) | 55 (24.8) | |
| >2 | 34 (15.8) | 22 (9.9) | |
| History of BV | 80 (37.2) | 54 (24.3) | |
| History of UTI | 150 (69.7) | 117 (52.7) | |
| Antecedent Yeast infections | 0 | 118 (54.8) | 141 (63.5) |
| 1–4 | 62 (28.8) | 61 (27.5) | |
| ≥5 | 35 (16.2) | 19 (8.6) | |
| Post Yeast Infections | 0 | 58 (26.9) | 93 (41.9) |
| 1–4 | 76 (35.3) | 90 (40.5) | |
| ≥5 | 81 (37.6) | 39 (17.6) | |
| Allergenic exposures and relevant conditions and medications | |||
| Reported Antibiotic Use (>6 months) | 19 (8.8) | 13 (5.9) | |
| Any allergy 2 | 195 (90.7) | 181 (81.5) | |
| History of Hives | 86 (40) | 67 (30.2) | |
| History of Insect Stings 3 | 14 (6.5) | 8 (3.6) | |
| Seasonal allergies | 125 (58.1) | 111 (50.0) | |
| History of Autoimmune Disease 4 | 24 (11.1) | 15 (6.8) | |
| Positive History of Functional Somatic Syndromes 5 | 103 (47.9) | 44 (19.8) | |
| Antecedent Functional Somatic Syndromes | 61 (28.3) | 21 (9.5) | |
| Psychosocial Factors | |||
| History of mod/severe abuse | 125 (58.1) | 114 (51.4) | |
| Antecedent Anxiety | 68 (31.6) | 44 (19.8) | |
| Post Anxiety | 39 (18.1) | 31 (14.0) | |
| Antecedent Mood | 66 (30.7) | 50 (22.5) | |
| Post Mood | 49 (22.7) | 50 (22.5) | |
| Rumination (tertiles) | T1 | 56 (26.0) | 82 (36.9) |
| T2 | 63 (29.3) | 71 (32.0) | |
| T3 | 79 (36.7) | 59 (26.6) | |
| Microbiome Characteristics | |||
| Community State Type | CST1–2 | 113 (52.6) | 108 (48.7) |
| CST 3 | 34 (15.8) | 44 (19.8) | |
| CST 4–6 | 68 (31.6) | 70 (31.5) |
Gynecological Infections: Total number of gynecological infections (gonorrhea, genital warts, bacterial vaginosis, trichomoniasis, Pelvic Inflammatory Disease, chlamydia, genital herpes)
Any allergy: Seasonal, sinus infections, contact dermatitis, medications, food
History of Stings: Greater than one sting with moderate and severe reaction.
Autoimmune conditions: Sjogren’s Syndrome, Crohn’s disease or Ulcerative Colitis, Diabetes, Multiple Sclerosis, Alopecia Areata, Celiac disease, Scleroderma, Systemic Lupus Erythematosus, Rheumatoid Arthritis
Functional Somatic Syndromes: Interstitial Cystitis, Chronic Fatigue Syndrome, Temporomandibular Joint (TMJ) disorder, Irritable Bowel Syndrome, Fibromyalgia
Culture based analyses of the vaginal microbiome showed no meaningful differences between cases and controls (see Supplemental Table S1). At the time of specimen collection, cases and controls had similar numbers of neutrophils and Nugent scores in vaginal smears. Quantitative vaginal culture results showed no differences between cases and controls for H202 negative or positive Lactobacillus species, Gardnerella vaginalis, Enterococcus, Escherichia coli, viridans group or Group B streptococci, and a number of anaerobic organisms. No differences were seen by culture-based methods when stratified by Shannon diversity. All confidence intervals shown in Table S1 include 1.0.
The vaginal microbiome Shannon Alpha Diversity was similar between cases and controls (see supplemental Figure S1) as was the distribution of community state types, shown in Table 1. However, we observed effect measure modification of associations for several risk factors for vulvodynia relative to comparable age periods among controls, by whether women had a vaginal microbiome with Shannon Alpha Diversity below the median (low) or above the median (high) (Figure 1). Among those with low diversity, but not among those with high diversity, there was a strong association between vulvodynia and increasing numbers of yeast infections prior to vulvodynia onset, history of moderate to severe childhood abuse, antecedent (prior to vulvar pain onset or comparable time period among controls) anxiety, depression and high levels of rumination with odds ratios ranging from 1.83 to 2.81 (Figure 1). The association with yeast infections remained strong after adjustment for psychosocial risk factors (Table S2). We repeated this analysis by measures of evenness and richness. Similar to the effect modification observed by alpha diversity with the associations with antecedent yeast infections, there was also effect modification by evenness. This was not true for richness (Figures S3 and S4). This suggests that the associations with alpha diversity were mainly attributable to uneven distributions of taxa.
Figure 1.
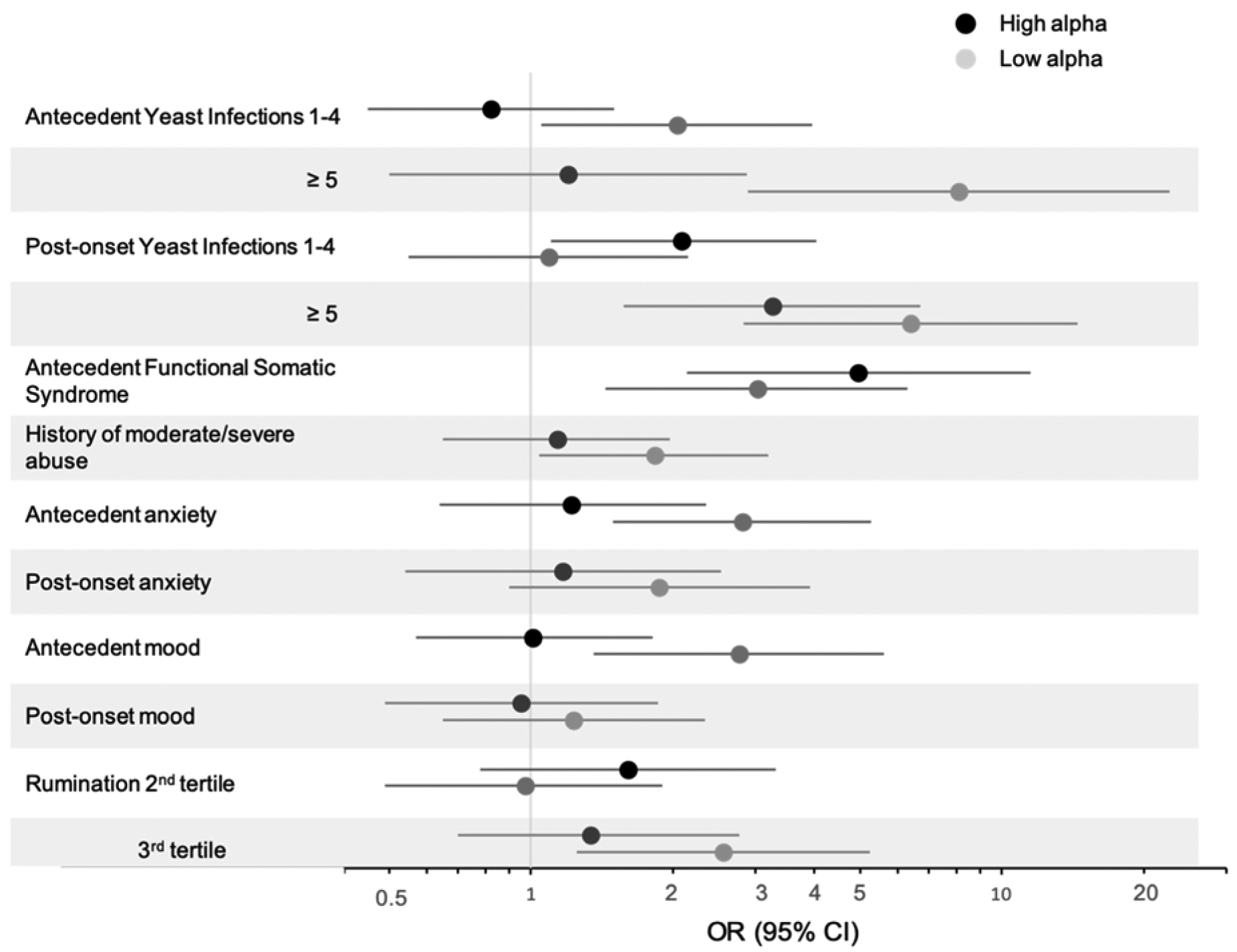
Age-adjusted odds ratios for yeast infections and psychosocial factors stratified by alpha diversity. Antecedent and post-onset categories defined as before or after the onset of vulvodynia in cases and before or after reference age in controls. Vulvodynia cases (n=215) and matched controls (n=222), 2010–2015.
The associations between psychological factors and vulvodynia among women with low alpha diversity microbiomes remained strong after adjustment for antecedent yeast infections and history of urinary tract infections (Table S3). Of note, a history of other co-morbid pain conditions was strongly associated with vulvodynia irrespective of alpha diversity. Also, the risk of yeast infections subsequent to vulvodynia onset and risk of psychiatric conditions subsequent to vulvodynia onset did not vary by alpha diversity in comparison to these risk factors prior to the onset of vulvar pain (or comparable time period among controls). We observed no effect measure modification by alpha diversity when we assessed the associations between history of allergies, medications, and medical conditions and vulvodynia.
We next stratified analyses by microbiome community state type (CST) membership assigned using Dirichlet multinomial mixed models, shown in a waffle plot (Figure 2). For these CST analyses, CST groups 4,5 and 6 were combined into one group due to low numbers and because they represented a mix of taxa that dominated the microbiome, but were less diverse than CST3. In addition, when analyses were carried out separately for CST4,5 and 6, we saw no differences in their modifying influences on the associations between antecedent or post-onset yeast infections, history of anxiety, or history of mood disorder and vulvodynia. CST1 and CST2 were kept separate as both groups are dominated by a single Lactobacillus species. CST1 was most closely related to Lactobacillus crispatus. CST2 was identified as L. iners. The association with antecedent yeast infections were largely confined to women with CST1 and 2 (Figure 3). There did not appear to be a specific CST associated with anxiety, but CST2 was more aligned with seeing an association between antecedent depression and vulvodynia.
Figure 3:
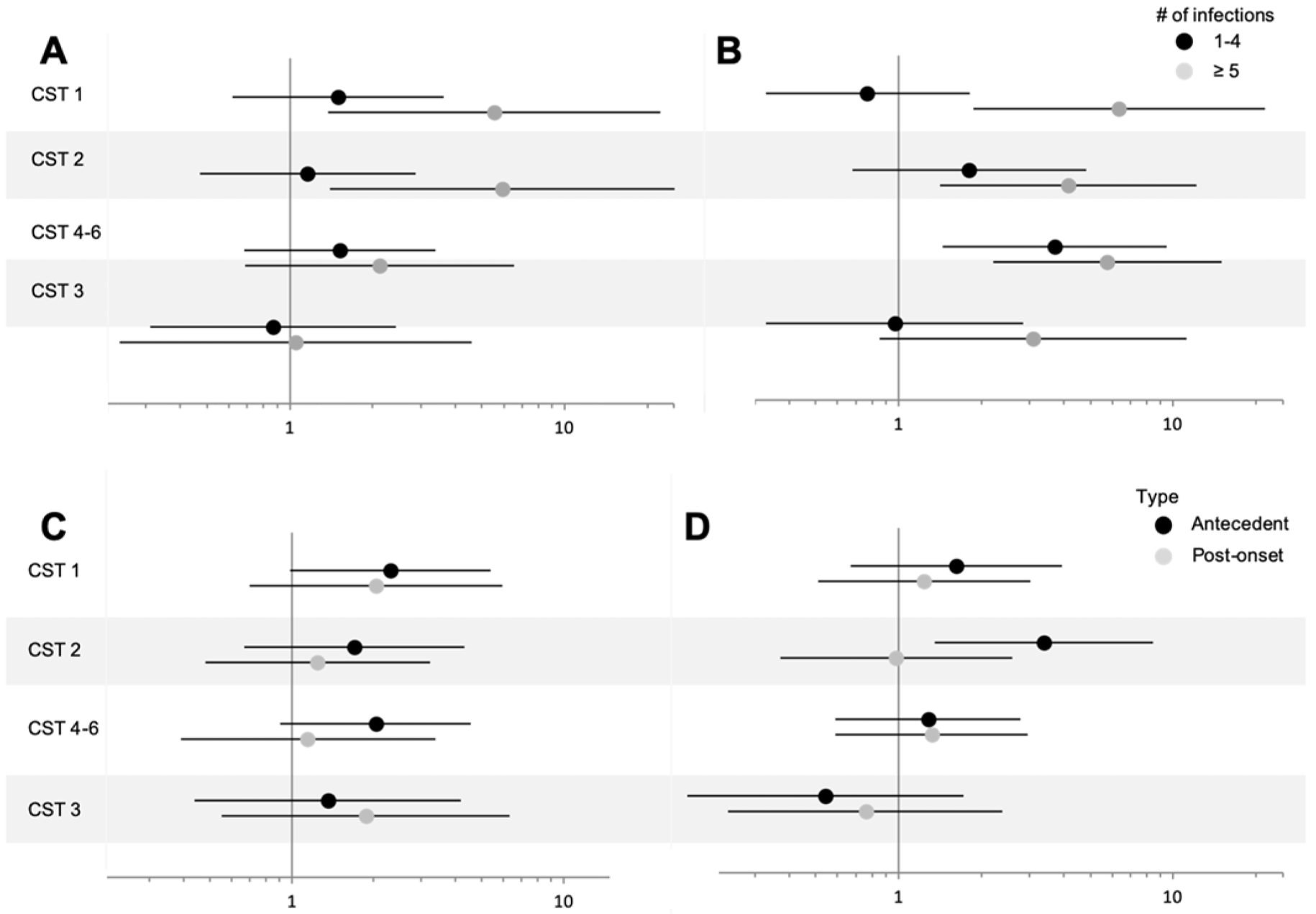
Age-adjusted odds ratios and 95% confidence intervals for antecedent yeast infections (A), post-onset yeast infections (B), history of anxiety (C), and history of mood disorder (D). For the yeast infections, zero was used as the reference. CST=community state type. Vulvodynia cases (n=215) and matched controls (n=222), 2010–2015.
Lastly, analysis using ALDEx2 (22; Appendix S1) showed no statistically significant associations of individual taxa abundance by vulvodynia status or community state type.
DISCUSSION
Principal Findings:
There was no overall microbiome differences between cases and controls. However, we observed associations between vulvodynia and history of a) antecedent yeast infections, and b) psychosocial risk factors such as history of childhood abuse, antecedent anxiety, mood disorders, and higher levels of rumination only among women with vaginal microbiomes that have alpha diversity below the median (low) but not alpha diversity above the median (high) as measured using the Shannon Diversity Index, an index that takes into account number of species and their relative abundance. Because the microbiome was measured at the time of vulvodynia and a comparable time period in controls, it is uncertain whether vaginal microbiome diversity modifies the association between these risk factors and vulvodynia, or becomes altered as a consequence of these associations.
Disease states such as bacterial vaginosis (BV) are associated with higher diversity of the vaginal microbiome (23). However, vaginal diversity was not associated with vulvodynia, but instead modified the associations between vulvodynia and previously studied risk factors. Most studies to date have focused on the impact of infectious diseases on the microbiome and are not comparable to the study of a multifactorial disease such as vulvodynia (24–26). The study by Jayaram et al (11) identified L. iners to be more prevalent and abundant in women with VVS whereas in controls L. crispatus was more prevalent and abundant. The study by Vadala et al (12) reported less lactobacillus species in cases compared to controls. However, neither study directly evaluated alpha diversity or community state types.
Clinical Implications:
It is too soon to speculate on the clinical implications of these findings. Women with vulvodynia are likely having less frequent sexual activity which has been suggested to be associated with a less diverse vaginal microbiome (27). However, we did not observe an association between microbiome diversity and vulvodynia. Instead, we saw that chronic yeast infections and psychosocial risk factors were strongly associated with vulvodynia only among women with less diverse vaginal microbiomes. Our data suggest that a more diverse microbiome environment may mitigate the deleterious effects of chronic infections and psychosocial morbidity shown to be associated with vulvodynia.
Research Implications:
Alpha diversity captures the number of species present and their relative abundance and is related to – but does not give specific information on – the emergent properties of a microbial community, such as stability (28). Thus, the modifying effect of diversity on psychosocial risk factors should be seen as a starting point rather than an explanation per se (28). Both high and low diversity populations have been associated with stability, and healthy and diseased states. For example, over the course of a healthy pregnancy, the vaginal microbiota becomes less diverse (29). This is thought to optimize colonization of the infant during labor and delivery, but as these less diverse communities tend to be predominated by acid producing bacteria, e.g. Lactobacillus species, it might also limit growth of pathogens (29). By contrast, bacterial vaginosis is characterized by a highly diverse vaginal microbial community and a higher pH. A limitation of our measure of diversity, based on sequencing data, is that it does not measure the absolute abundance of taxa present. That is, the same diversity can be present with dense biomass, as might be found in women with and without bacterial vaginosis. However, the quantity of selected organisms identified using culture techniques (expressed as log10 colony forming units per gram of vaginal secretions) did not differ among those with low and high diversity microbiomes.
As there were no single taxa or groups of taxa associated with vulvodynia our findings should stimulate us to look for and identify factors that might decrease vaginal diversity. Several hypotheses come to mind. First, women with repeated yeast infections also are more likely to have atopic diseases – eczema, allergic rhinitis and asthma – suggesting heightened local innate immunity which may result in changes in the vaginal microbiota (30). Candida species are also human commensals; the majority of women are colonized with Candida species at one or more times in their lives usually without symptoms (31). Thus, Candida species may play an important role in the development and maintenance of normal mixed species biofilms found in the vaginal cavity (31). Second, women with multiple yeast infections were most likely treated with multiple courses of antifungals, and they often follow urinary tract infections, which are treated with antibiotics. Antifungals and antibiotic treatment can disrupt the vaginal microbiota (31). Third, the change in diversity may be mediated by infection with a bacteriophage (viruses that infect bacteria) known to be important mediators of microbial communities in other systems (32).
Strengths and Limitations:
Our study used clinically confirmed cases of vulvodynia and general population controls. Additionally, we assessed temporality with respect to the explored risk factors using interview questions targeted to before and after onset of vulvodynia (cases) or a comparable reference age (controls). A further strength is the collection of orthogonal data including both standard culture-based microbiological methods and newer sequencing-based methods for quantitation of bacterial taxa.
This study is limited in its interpretability due to the lack of, and timing of the vaginal microbiome sample. The vaginal community was assessed at the time of vulvodynia and probably reflects a combination of response to the disease process and the ongoing processes that maintain symptoms. Since vaginal samples were taken after the onset of vulvodynia, we cannot make conclusions on the temporality of associations seen, particularly with respect to the onset of vulvodynia. Participants were asked to recall the onset of certain conditions as either before or after vulvar pain onset or reference age, which could result in recall bias. However, Harlow et al (8) used quantitative bias analyses to evaluate the possibility of recall bias in this population and suggested that the observed associations for antecedent yeast infections were likely an underestimate of the true effect, except under less plausible scenarios of perfect specificity and no false positive yeast infections among women with no history of vulvar pain. Additionally, this study was conducted in premenopausal women 18–40 years of age who were largely restricted to those self-reported as Caucasian. Thus, we could not evaluate the contribution of these findings to postmenopausal women, or to ethnic diversity which has been shown to be a factor influencing the vaginal microbiome (33). Lastly, the size of our dataset did not allow us to assess more than a dichotomous level of effect measure modification by microbiome diversity. Nevertheless, we pose a hypothesis that we hope others will assess within larger and more temporally appropriate datasets.
Conclusions:
We evaluated the vaginal microbiome in a large, well-characterized population of women with and without vulvodynia and found no overall differences. However, we found strong and significant associations between vulvodynia and known risk factors (e.g. yeast infections, childhood abuse, anxiety, and mood disorders) in women with low diversity vaginal microbiomes, and these associations were not attenuated when controlling for other risk factors.
Supplementary Material
AJOG at a Glance:
-
Why was the study conducted?
Vulvodynia affects 3–7% of reproductive aged women and its etiology is relatively undefined.
-
What are the key findings?
Characteristics of the vaginal microbiome do not differ between women with and without vulvodynia. However, known associations between certain risk factors and vulvodynia differ by the diversity of the vaginal microbiome.
-
What does this study add to what is already known?
Although our study supports a previous finding that characteristics of the vaginal microbiome are not associated with vulvodynia, our novel results suggest that the diversity of the vaginal microbiome may promote or reflect an environment that enables other factors to influence the risk of vulvodynia.
Funding:
National Vulvodynia Association and NIH-NICHD R01 HD058608.
Author Disclosure Statement: No competing financial interests exist.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Details of Ethics Approval: This study was approved by the Human Subjects Research Committee at the University of Minnesota. Written informed consent was obtained from all participating women. University of Minnesota IRB most recent approval of Continuing Review was January 2, 2019. Main study approval was September 29, 2009.
References:
- 1.Harlow BL, Kunitz CG, Nguyen RHN, Rydell SA, Turner RM, MacLehose RF. Prevalence of symptoms consistent with a diagnosis of vulvodynia: population-based estimates from 2 geographic regions. Am J Obstet Gynecol. 2014; 210(1)40.e1–40.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnold LD, Bachmann GA, Rosen R, Rhoads, GG. Assessment of vulvodynia symptoms in a sample of US women: a prevalence survey with a nested case control study. Am J Obstet Gynecol. 2007;196(2):128.e1–128.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pukall CF, Goldstein AT, Bergeron S, Foster D, Stein A, Kellog-Spadt S, Bachmann G. Vulvodynia: Definition, Prevalence, Impact and Pathophysiological Factors. Sexual Medicine. 2016;13:291–304. [DOI] [PubMed] [Google Scholar]
- 4.Havemann LM, Cool DR, Gagneux P, Markey MP, Yaklic JL, Maxwell RA, et al. Vulvodynia: What We Know and Where We Should Be Going. J Lower Gen Tract Dis. 2017;21(2):1–7. [DOI] [PubMed] [Google Scholar]
- 5.Foster DC and Hasday JD. Elevated tissue levels of interleukin-1 beta and tumor necrosis factor0alpha in vulvar vestibulitis. Obstetrics and gynecology. 1997;89(2):291–6. [DOI] [PubMed] [Google Scholar]
- 6.Gerber S, Bongiovanni AM, Ledger WJ, Witkin SS. Interleukin-1β gene polymorphism in women with vulvar vestibulitis syndrome. Eur J Obstet Gynecol Reprod Biol. 2003;107:74–77. [DOI] [PubMed] [Google Scholar]
- 7.Harlow BL, He W, Nguyen RHN. Allergic reactions and risk of vulvodynia. Ann Epidemiol. 2009;19:771–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harlow BL, Caron RE, Parker SE, Chatterjea D, Fox MP, Nguyen RHN. Recurrent Yeast Infections and Vulvodynia: Can we believe associations based on self-reported data? J Women’s Health. 2017;26(10):1069–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campisciano G, Zanotta N, Licastro D, De Seta F, and Comar M. In vivo microbiome and associated immune markers: New insights into the pathogenesis of vaginal dysbiosis. Sci Rep. 2018;8:2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gosmann C, Anahtar MN, Handley SA, Farcasanu M, Abu-Ali G, Bowman BA, et al. Lactobacillus-deficient cervicovaginal bacterial communities are associated with increased HIV acquisition in young South African Women. Immunity. 2017;46(1): 29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jayaram A, Witkin SS, Zhou X, Brown CJ, Rey GE, Linhares IM, et al. The bacterial microbiome in paired vaginal and vestibular samples from women with vulvar vestibulitis syndrome. Pathogens and Disease. 2014; 72: 161–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vadala M, Testa C, Coda L, Angioletti S, Guiberti R, Laurino C, Palmieri B. Vulvovestibular syndrome and vaginal microbiome: A simple evaluation. J Clin Med Res 2018;10:688–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bornstein J, Goldstein AT, Stockdale CK, Bergeron S, Pukall C, Zolnoun D, et al. ISSVD, ISSWSH, and IPPS consensus terminology and classification of persistent vulvar pain and vulvodynia. J Lower Gen Tract Dis. 2016;20:1–5. [DOI] [PubMed] [Google Scholar]
- 14.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. “Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform .” Applied and Environmental Microbiology. 2013;79(17):5112–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eren AM, Morrison HG, Lescault PJ, Reveillaud J, Vineis JH, & Sogin ML (2015). Minimum entropy decomposition: unsupervised oligotyping for sensitive partitioning of high-throughput marker gene sequences. The ISME journal, 9(4), 968–979. doi: 10.1038/ismej.2014.195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eren AM, Maignien L, Sul WJ, Murphy LG, Grim SL, Morrison HG, & Sogin ML (2013). Oligotyping: Differentiating between closely related microbial taxa using 16S rRNA gene data. Methods in ecology and evolution, 4(12), 1111–1119. doi: 10.1111/2041-210X.12114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA and Holmes SP (2016). “DADA2: High-resolution sample inference from Illumina amplicon data.” _Nature Methods_, *13*, pp. 581–583. doi: 10.1038/nmeth.3869 (URL: http://doi.org/10.1038/nmeth.3869). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holmes I, Harris K, Quince C. “Dirichlet multinomial mixtures: generative models for microbial metagenomics”. PloS One. 2012; 7:e30126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.R Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing, 2017. Available at: https://www.R-project.org/. [Google Scholar]
- 20.Morgan M. DirichletMultinomial: Dirichlet-Multinomial Mixture Model Machine Learning for Microbiome Data. 2017.
- 21.Pearce N. Analysis of matched case control studies. British Med J 2016;352:i969 | doi: 10.1136/bmj.i969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fernandes AD, Macklaim JM, Linn TG, Reid G, Gloor GB. “ANOVA-Like Differential Gene Expression Analysis of Single-Organism and Meta-RNA-Seq.” PLoS One. 2013;8(7):e67019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greenbaum S, Greenbaum G, Moran-Gilad J, and Weintraub AY. Ecological dynamics of the vaginal microbiome in relation to health and disease. AJOG. 2019: 324–335. [DOI] [PubMed] [Google Scholar]
- 24.Fredricks DN, Fiedler TL, and Marrazzo JM. Molecular Identification of Bacteria Associated with Bacterial Vaginosis. NEJM. 2005; 353: 1899–911. [DOI] [PubMed] [Google Scholar]
- 25.Lee JE, et al. Association of the vaginal microbiota with human papillomavirus infection in a Korean twin cohort. PLoS One 2013; 8(5): e63514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Borgdorff H, Tsivtsivadze E, Verhelst R, Marzorati M, Jurriaans S, Ndayisaba GF, et al. Lactobacillus-dominated cervicovaginal microbiota associated with reduced HIV/STI prevalence and genital HIV viral load in African women. ISME. 2014; 8: 1781–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lewis FM, Bernstein KT, Aral SO. Vaginal microbiome and its relationship to behavior, sexual health, and sexually transmitted diseases. Obstet Gynecol 2017;129:643–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shade A. Diversity is the question, not the answer. ISME. 2017; 11: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacIntyre DA, Chandiramani M, Lee YS, Kindinger L, Smith A, Angelopoulous N. The vaginal microbiome during pregnancy and the postpartum period in a European population. Sci Rep. 2015; 5: 8988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sobel JD. Recurrent vulvovaginal candidiasis. Am J Obstet Gynecol. 2016; 214: 15–21. [DOI] [PubMed] [Google Scholar]
- 31.Goncalves B, Ferreira C, Alves CT, Henriques M, Azeredo J, and Silva S. Vulvovaginal candidiasis: Epidemiology, microbiology and risk factors. Crit Rev Microbiol. 2016; 42(6): 905–27. [DOI] [PubMed] [Google Scholar]
- 32.Maslov S and Sneppen K. Population cycles and species diversity in dynamic Kill-the-Winner model of microbial ecosystems. Sci Rep. 2017; 7: 39642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ravel J, Gajera P, Abdob Z, Schneiderc GM, Koeniga SSK, McCullea SL, et al. Vaginal microbiome of reproductive-age women. PNAS. 2011;108(1):4680–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


