Fig. 3. TR regulation of astrocyte reactivity DEGs, chromatin accessibility and disorder outcome.
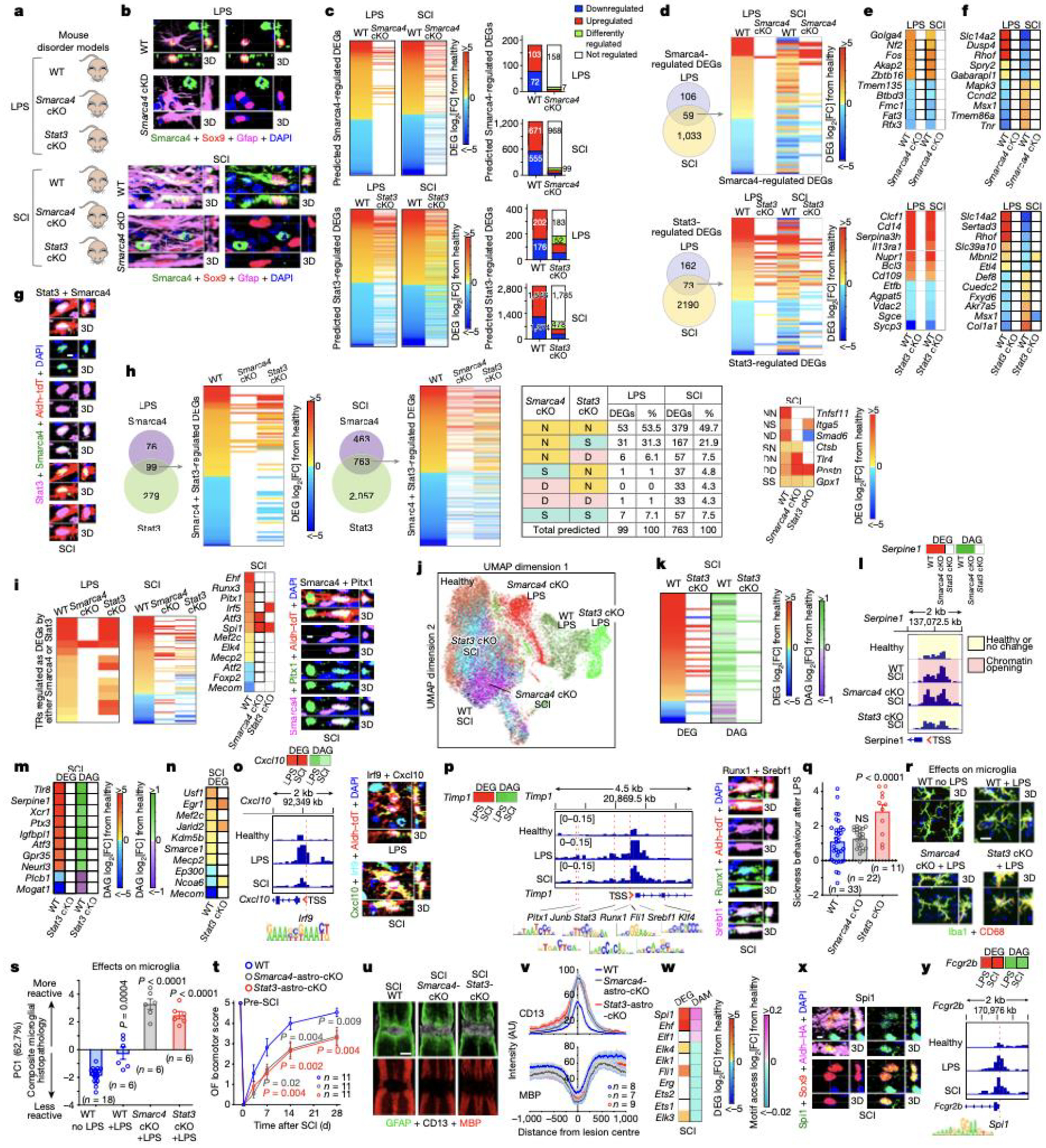
a. Experimental models. b. Smarca4 immunohistochemistry. c. Smarca4-astro-cKO or Stat3-astro-cKO effects on Smarca4- or Stat3-regulated genes. d. TREA-predicted Smarca4- or Stat3-regulated DEGs in both LPS and SCI, and astro-cKO effects. e,f. DEGs regulated in the same (e) or in opposite (f) directions by Smarca4 or Stat3. g. Stat3 and Smarca4 in same reactive astrocyte. h. TREA-predicted Smarca4- and Stat3-regulated DEGs and astro-cKO effects on those DEGs. i. TRs significantly regulated as DEGs; Smarca4 and Pitx1 in same reactive astrocyte. j. UMAP clustering of astrocyte nuclei from WT, Smarca4-astro-cKO or Stat3-astro-cKO mice based on chromatin accessibility after LPS or SCI. k. Stat3-cKO effects on DEG and DAG. l. Smarca4-astro-cKO or Stat3-astro-cKO effects on Serpine1 DEG, DAG; and ATAC peaks in Serpine1 TSS. m. Stat3-astro-cKO effects on DEG and DAG of 10 genes without Stat3 DNA-binding motifs. n. Stat3-astro-cKO effects on 10 chromatin regulators. o. Cxcl10 DEG and DAG; ATAC peaks in Cxcl10 TSS and Irf9 DNA-binding motif; Irf9 and Cxcl10 in same reactive astrocyte. p. Timp1 DEG and DAG; ATAC peaks in Timp1 TSS and DNA-binding motifs of 7 predicted TRs; Runx1 and Srebf1 in same reactive astrocyte. q. Sickness behavior after LPS. r. Microglia immunohistochemistry after LPS. s. PCA of composite histopathology after LPS. t. Open field (OF) locomotor recovery after SCI. u,v. Images and graph of mean±sem staining intensity for Gfap, CD13 and myelin basic protein (MBP). w. ETS family TRs DEG and differential motif access (DAM) after SCI. x. PU.1 (Spi1) in reactive astrocytes. y. Fcgr2b DEG and DAG; ATAC peaks in Fcgr2b TSS and PU.1 DNA-binding motif. Quantitative values in q,s,t are mean ± sem analyzed by one-way (q,s) or two-way (t) ANOVA with Bonferroni’s test, P values are cKO versus WT LPS or SCI, NS non-significant. n = mice.
