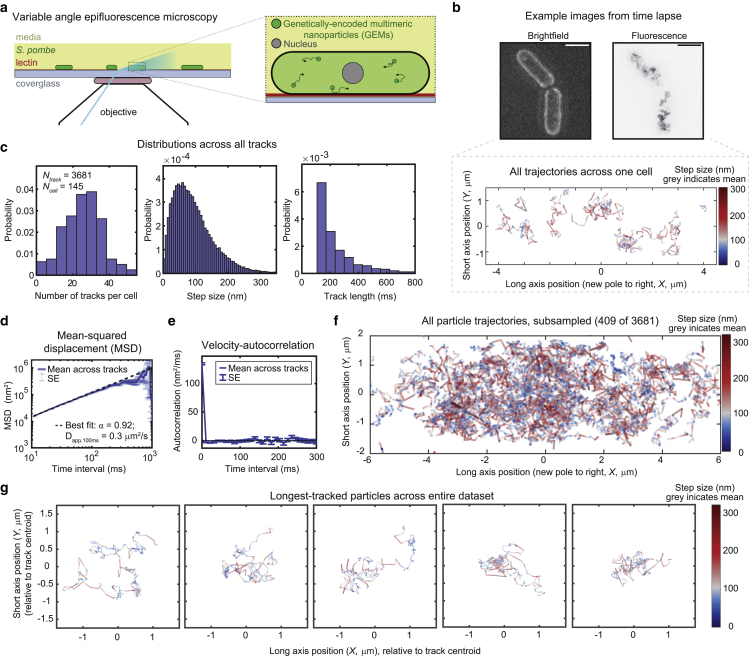
Figure 1.
High-speed particle-tracking nanorheology of GEMs allows detailed statistical analysis of cytoplasmic diffusion. (a) Schematic of the experimental imaging set-up. (b) Example bright-field image (top left) and maximum intensity projection through time of the GEM particle fluorescence (top right) for one representative field of view, alongside the measured nanoparticle trajectories (bottom) for the upper cell in the image. Trajectories are colored by the step size of the particle in nanometers between each time frame of the movie. Gray indicates the mean step size across all tracks in the data set. Scale bar, 5 μm. (c) Histograms of the number of tracks per cell (left), the step size for all time points (middle), and the duration of time that each particle was tracked (right). Note that tracks shorter than 10 time points were not included in the analysis. (d) The mean-squared displacement (MSD) of the particle tracks. The time-averaged MSD was first calculated individually for each track, and then a second averaging was performed to find the (ensemble averaged) MSD across all tracks. Note the logarithmic scale along the x and y axes. (e) The average velocity autocorrelation across all particle tracks. Averaging was performed in the same order as the MSD. (d and e) Error bars represent the standard error (SE). (f) Plots of particle trajectories drawn from many experiments and cells, randomly subsampled for better visibility of individual particle behaviors. Subsampled trajectories include at least one track from 141 of the 145 cells in the data set. Gray indicates the mean step size across all tracks in the data set. (c–f) The data set includes 3681 tracks among 145 cells, recorded from 5 different samples and over 3 different days. (g) Individual trajectory plots for five of the longest-tracked particles (in time), excluding stationary particles. Color scaling of the step size was identical in all panels included in (f and g) (using the mean and SD of the step size across the entire data set).