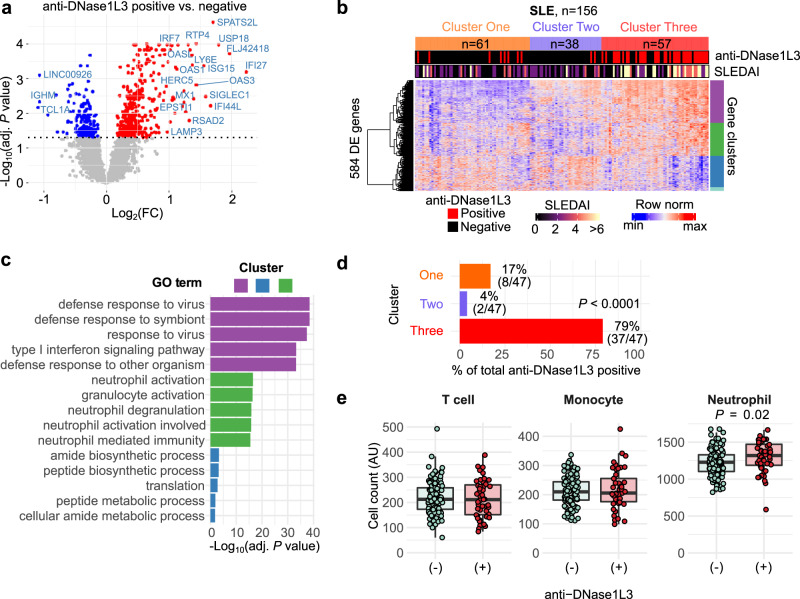
Fig. 2. Transcriptional correlates of anti-DNase1L3 antibodies in SLE.
a Volcano plot of 584 differentially expressed transcripts (DETs) between SLE patients positive and negative for anti-DNase1L3 antibodies. Red, 399 upregulated DETs with adjusted P < 0.01. Blue, 185 downregulated DETs with adjusted P < 0.01. DETs with P value < 0.01 and Log2(Fold change) >1 are marked (b). Hierarchical clustering of 584 DETs from a. Each column represents an individual patient and each row an individual gene. Top annotations show cluster membership, anti-DNase1L3 antibodies (positive = red, negative = black), and SLEDAI score in continuous scale. DETs were split in k = 4 expression clusters and annotated by functional enrichment analysis using the g: Profiler toolset with the gene oncology molecular function (GO:MF) gene set collection. Red represents upregulated genes and blue downregulated genes. Only 156/158 of SLE patients from Fig. 1a had paired microarray and serum data. SLE patients clustered in three major groups defined by upregulation of genes related to mRNA processing and translation (Cluster One), upregulation of mRNA processing, translation, and some IFN-stimulated genes (ISGs) (Cluster Two), and upregulation of ISGs and neutrophil activation genes (Cluster Three). c Top 5 enriched GO:MF terms on gene expression clusters according to P value. d Frequency of anti-DNase1L3 antibody positive SLE patients according to cluster membership in b. e Comparison of the MCP counter deconvolution score for T-cells, monocytes and neutrophils, between anti-DNAse1L3 positive (n = 47) vs. anti-DNAse1L3 negative (n = 109) SLE. GO gene ontology, FC fold-change.