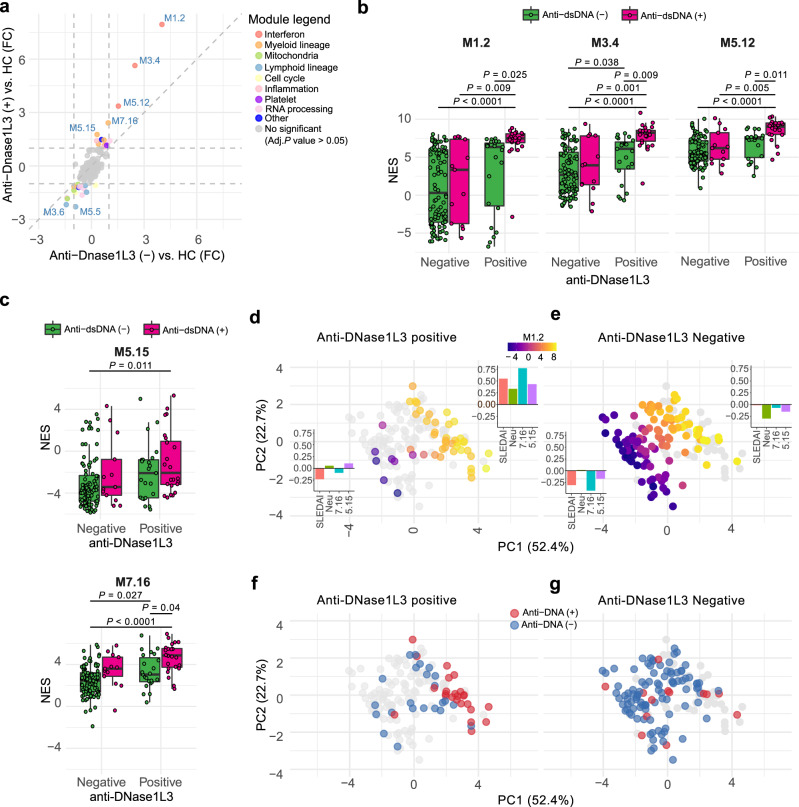
Fig. 3. Transcriptional fingerprints associated with anti-DNase1L3 antibodies in SLE.
a Four-way plot of the differentially regulated blood expression modules between anti-DNase1L3 (+) SLE vs. healthy controls (HC) (data from Supplementary Table 2), and anti-DNase1L3 (−) SLE vs. HC (data from Supplementary Table 3). FC, Fold change. b, c Comparison of the ssGSEA normalized enrichment scores (NES) for the IFN modules M1.2, M3.4, and M5.12 (b), and the myeloid lineage modules M5.15 and M7.16 (c) in SLE patients according to anti-DNase1L3 antibody positivity. Variables were compared using two-way ANOVA with Tukey’s test as post-hoc. d, e PCA projection of the significantly regulated modules (M1.2, M3.4, M5.2, M5.15, and M7.16), neutrophil count, and disease activity in SLE subjects according to anti-DNase1L3 antibody positive (d) or anti-DNase1L3 negative (e) status. Color scale represents the activity of the IFN module M1.2 (ssGSEA NES score). Bar graphs represent the mean Z score of SLEDAI, neutrophil count (Neu) and activity of the modules M5.15 and M7.16 of patients according to high or low M1.2 activity, defined as patients with PC1 > 0 or PC1 ≤ 0, respectively. f Distribution of anti-DNA antibody positive or negative status of anti-DNase1L3 positive SLE patients in PCA projection from d. g Distribution of anti-DNA antibody positive or negative status of anti-DNase1L3 negative SLE patients in PCA projection from e.