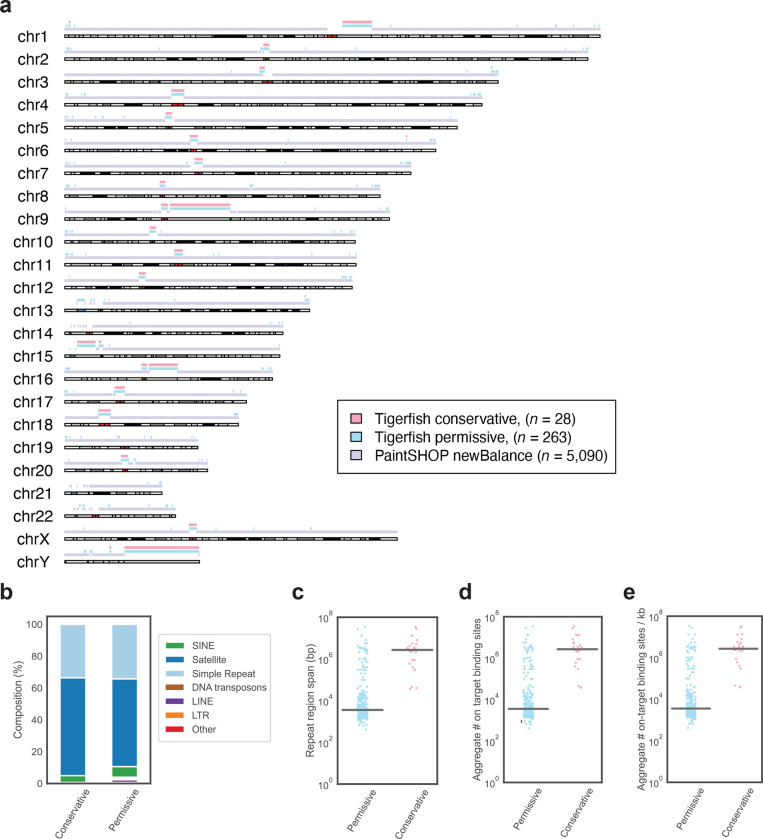
Fig. 2 |. Genome-scale probe design with Tigerfish.
a, Schematic visualization of intervals for which Tigerfish probe sets were identified using conservative (pink) or permissive (teal) parameters and intervals covered by existing PaintSHOP probes designed using parameters suitable for non-repetitive targets (lilac). b, The distribution of RepeatMasker annotations for intervals identified and processed by Tigerfish using conservative and permissive settings. c, Length distributions of the regions identified and targeted by Tigerfish using conservative and permissive parameters. d, The aggregate number on-target binding predictions for probe sets designed by Tigerfish using conservative and permissive parameters. e, The aggregate number on-target binding predictions per kilobase for probe sets designed by Tigerfish using conservative and permissive parameters.