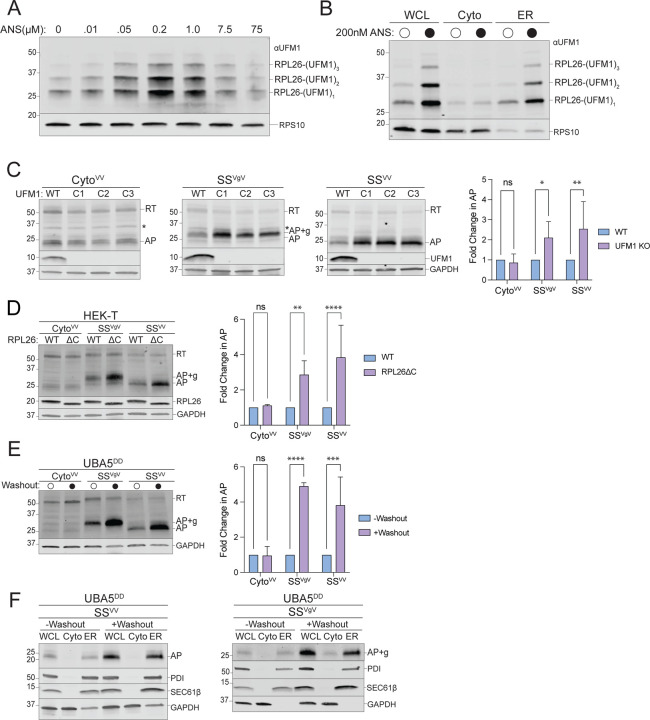
Figure 4: ER-AP degradation requires UFMylation.
A: Global ribosome stalling promotes RPL26 UFMylation. U2OS cells were treated with the indicated concentrations of anisomycin (ANS) for 20 min treatment prior to harvesting. Cell lysates were sedimented through a sucrose cushion and the pellets were analyzed by immunoblot using an anti-UFM1 antibody. RPS10: loading control.
B: Stalling induced RPL26 UFMylation occurs primarily on ER-bound ribosomes. U2OS cells were treated with either DMSO or 200 nM ANS for 20 min prior to cell fractionation and immunoblotting with UFM1. RPS10: loading control; data shown are representative of two independent experiments.
C: Knockout of UFM1 stabilizes ER- but not cytosolic-APs. Left panels, HEK293 WT and clonal UFM1KO cell lines (C1, C2, C3) were transfected with the indicated reporters. Reporter products were analyzed by immunoblot with FLAG antibody. Asterisks indicate a nonspecific immunoreactive band. Knockouts were confirmed by blotting with antibodies against endogenous UFM1 protein. GAPDH: loading control. Right panel, Quantification of AP intensity for CytoVV and SSVV and AP+g intensity for SSVgV. Fold change relative to WT cells was calculated after normalization to GAPDH. Data are the mean ± SD of at least 3 independent experiments. ns > 0.05, *p < 0.05, **p < 0.01, determined by two-way ANOVA.
D: Specific UFMylation of RPL26 is required for ER-AP degradation. Left panel, HEK-T WT and clonal RPL26ΔC cell lines were transfected with the indicated reporters. Reporter products were analyzed by immunoblot with FLAG antibody. C-terminal deletion of RPL26 was confirmed by blotting with an antibody against endogenous RPL26 protein. GAPDH: loading control. Right panel, Quantification of AP intensity for CytoVV and SSVV and AP+g intensity for SSVgV. Fold change relative to WT cells was calculated after normalization to GAPDH. Data are the mean ± SD of at least 3 independent experiments. ns > 0.05, **p < 0.01, ****p < 0.0001 determined by two-way ANOVA.
E: Acute disruption of UFMylation stabilizes ER- but not cytosolic-APs. Left panel, U2OS UBA5DD cells were cultured in complete DMEM with TMP (“-Washout”) or washed to remove TMP from the media (“+Washout”) before transfection with the indicated reporters. Reporter products were analyzed by immunoblot with FLAG antibody. GAPDH: loading control. Right panel, Quantification as in Fig 4C. Fold change relative to WT cells was calculated after normalization to GAPDH. Data are the mean ± SD of at least 3 independent experiments. ns > 0.05, ***p < 0.001, ****p < 0.0001, determined by two-way ANOVA.
F: Acute disruption of UFMylation favors release of ER-APs into ER lumen. As in Fig 4D, followed by cell fractionation. Reporter products were analyzed by immunoblot of WCL, Cyto, and ER cell fractions with FLAG antibody. GAPDH: cytosol marker; SEC61β and PDI: ER markers; data shown are representative of two independent experiments.