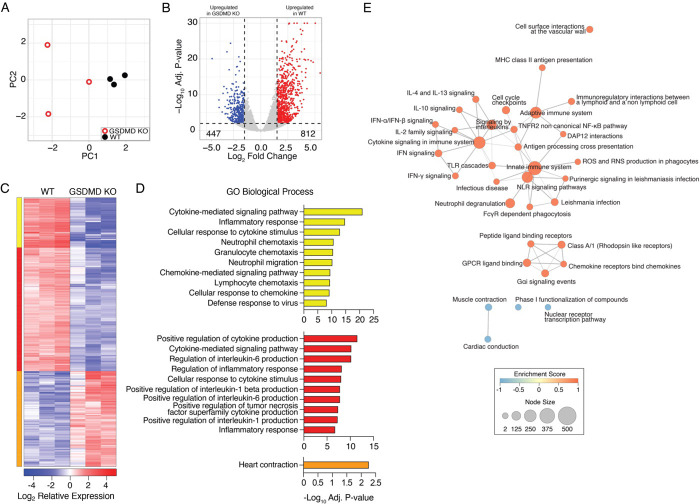
Figure 3: GSDMD promotes inflammatory gene expression programs.
WT and GSDMD KO mice were infected with IAV strain PR8 at a dose of 50 TCID50. A-E RNA was extracted from lungs at day 7 post infection and subjected to RNA sequencing. A Principal component analysis comparing WT and GSDMD KO lung RNA sequencing results. B Volcano plot of differential gene expression (lfc |1.5|, adj p-value <0.01) comparing WT vs GSDMD KOs. Red, 812 genes upregulated in WT vs KO; Blue, 447 genes downregulated in WT vs KO. C Hierarchical clustering of differentially expressed genes following IAV infection. Heatmap represents relative gene expression where red indicates genes with upregulated expression and blue indicates genes with downregulated expression. Cluster color indicates genes with similar patterns of expression in WT infected mice compared to GSDMD KO. D Gene ontology analysis for genes within each cluster. Bar graphs represent the top 10 enriched GO terms enriched within each cluster as indicated by color. Bar length represent the −log10 adj. p value for significantly enriched pathways (−Log10 adj p-value > 1.3). E Network of GO Biological Process terms enriched by gene set enrichment analysis (GSEA). Node size represents number of genes within each pathway. Edges represent the number of shared genes across GO BP terms. Color indicates the GSEA Enrichment Score (ES) where orange indicates positive ES and blue indicates negative ES for transcriptional signatures derived from WT infected lungs relative to GSDMD KO.