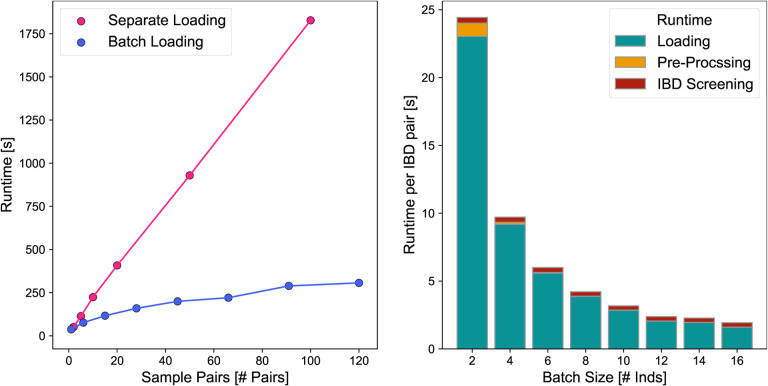
Figure E1: Run-time Benchmarks of ancIBD.
To benchmark run-times, we applied ancIBD on empirical ancient DNA data in .hdf5 format imputed at 1240k sites. We used the imputed hdf5 file from the Eurasian application (Fig. 3), choosing samples and pairs at random. Left: For each sample pair, all autosomes are screened for IBD. In one experiment all pairs of samples were run independently, leading to a linear dependency on pair number, as expected. In a second experiment, all samples were loaded into memory, and then each sample pair was screened for IBD. The apparent sub-linear behavior is due to the fact that loading n samples scales slower than the actual runtime of n·(n = 1)/2 sample pairs. Right: We depict the runtimes normalized per sample pair when screening all pairs of sample batches of various sizes for IBD. We visualize the loading time (the time it takes to load the hdf5 genotype data into memory), the pre-processing time (including preparing the transition and emission matrix), as well as the runtime of screening for IBD that includes the forward-backward algorithm as well as post-processing. Due to the decrease in the impact of the time to load the data, which scales linearly with batch size while the number of sample pair scales quadratically, we observe substantially increased runtimes per pair.