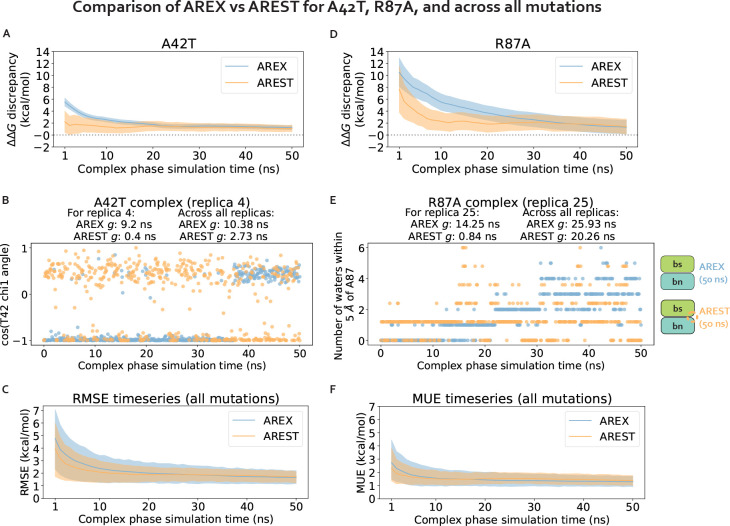
Figure 7. Alchemical replica exchange with solute tempering (AREST) and alchemical replica exchange (AREX) demonstrate comparable convergence for most barnase:barstar mutations.
(A) discrepancy (with respect to experiment) time series for A42T. The discrepancy was computed as , where corresponds to the (AREX or AREST) complex phase at a particular time point, corresponds to the apo phase computed from a 10 ns/replica AREX simulation, and is the experimental value from Schreiber et al [73]. AREX time series shown in blue and AREST time series (with radius = 0.5 nm, ) shown in orange. Number of states is 24 for both AREX and AREST. Shaded regions represent ± two standard deviations, computed by bootstrapping the decorrelated reduced potential matrices 200 times. Gray dashed line indicates discrepancy = 0. (B) Time series of the angle for residue T42 for a representative replica (replica 4) of the A42T complex phase AREX simulation (blue) and AREST simulation (orange) (number of states = 24, simulation time = 50 ns/replica). indicates statistical inefficiency, which is proportional to the correlation time. indicates very thorough sampling (because the sampling interval is 0.1 ns) and large values of indicate poor sampling. (C) Time series of the root mean square error (RMSE) (with respect to experiment) for the of all barnase:barstar mutations. The used to compute the RMSE at each time point were computed as for each mutation, where corresponds to the (AREX or AREST) complex phase at a particular time point and corresponds to the apo phase computed from a 10 ns/replica AREX simulation. AREX time series shown in blue and AREST time series (with radius = 0.5 nm, ) shown in orange. Number of states is 24 for neutral mutations and 36 for charge-changing mutations. Shaded regions represent ± two standard deviations, computed by bootstrapping 1000 times. (D) Same as (A), but for R87A instead of A42T. Number of states is 36 for both AREX and AREST. (E) Time series of the number of waters within 5 Å of residue A87 for representative replica (replica 25) of the R87A complex phase AREX simulation (blue) and AREST simulation (orange) (number of states = 36, simulation time = 50 ns/replica). (F) Same as (C) but for mean unsigned error (MUE) instead of RMSE.