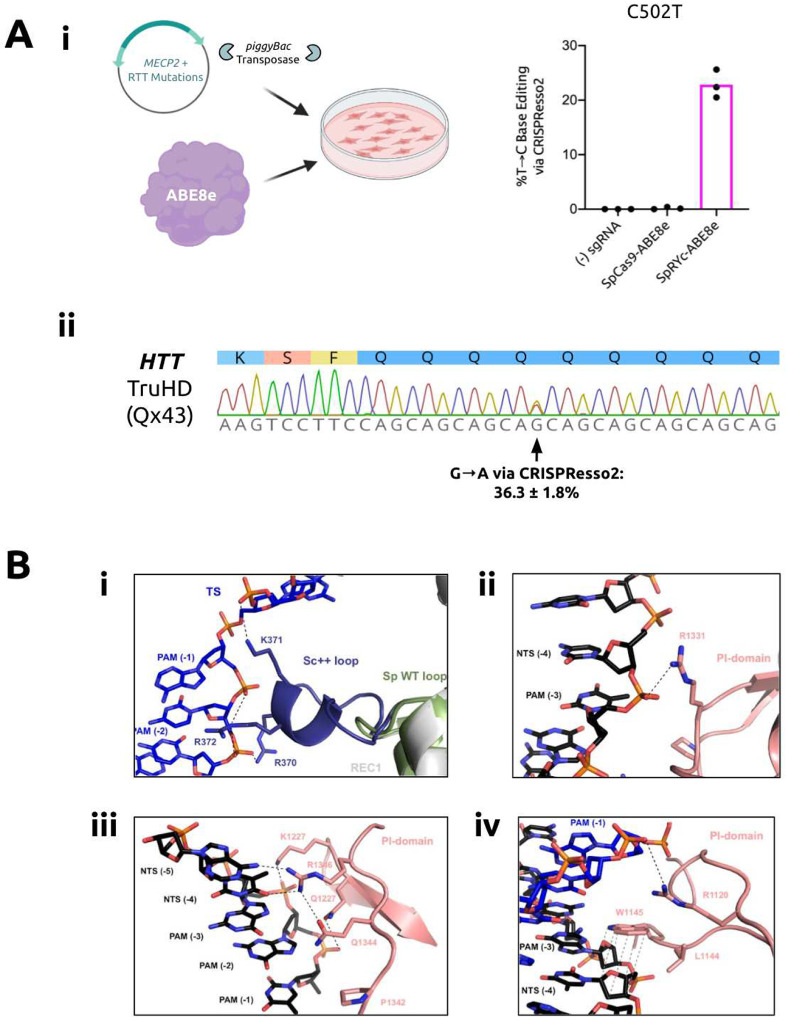
Figure 3. Potential applications and structural mechanisms of SpRYc.
(A) Targeting disease-associated loci with SpRYc. (i) Schematic of SpRYc RTT Experiment. Base editing conversion rates were determined via CRISPResso2 NGS analysis following PCR amplification of MECP2-integrated loci, in comparison to unedited controls for the C502T installed mutation. Samples were performed in independent nucleofection triplicates (n=3) and the mean of the quantified base editing formation values was calculated. (ii) SpRYc-BE4Max was nucleofected into TruHD cells alongside an sgRNA targeting the HTT repeat. Base editing conversion rate was determined via CRISPResso2 NGS analysis NGS following PCR amplification of indicated genomic loci, in comparison to an unedited control. The analogous Sanger sequencing trace is shown. Samples were performed in independent nucleofection triplicates (n=3) and the mean of the quantified base editing formation values was calculated. (B) Structural insights via homology modeling in SWISS-MODEL. (i) Interaction of the engineered Sc++ loop (purple) with the backbone of the target strand (TS) PAM region. The REC1 loop from wild type SpCas9 is indicated in green. (ii) Potential interaction of residue R1331 with the non-target strand (NTS) backbone. (iii) Multiple mutations within the PAM interaction loop allow for a more flexible PAM readout. (iv) The potential van der Waals interaction of W1145 with the ribose moieties of non-target strand residues could further stabilize the PAM interaction.