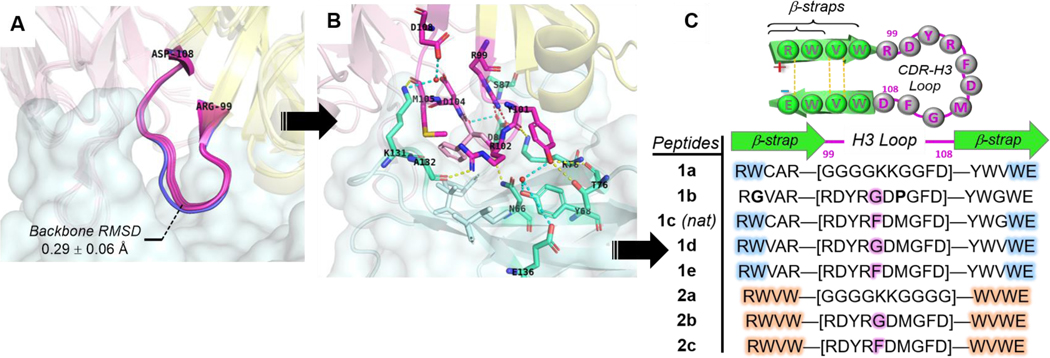
Figure 3.
Rational design of pembrolizumab H3 loop mimics. Comparison of binding modes from. (A) Overlay of H3 loops [RH99 – DH108] excised from co-crystal structures (PDB: 5JXE, 5B8C, 5GGS, in magenta) with its unbounded form (PDB: 5DK3, in blue). (B) Close-up view of pembrolizumab CDR-H3 contact interactions at the interface with PD1; Ribbon representation of the binding complex (PDB code: 5GGS) with PD1 in cyan, and the heavy and light chains of pembrolizumab in yellow and pink respectively. The network of H-bonds and hydrophobic contacts from PD1 residues depicted in the same lighter colors. Direct and water mediated H-bonds are displayed with yellow and cyan dashed lines respectively, and one salt bridge with a red dashed line. (C) Summary of peptide H3-mimics 1a-e and 2a-c prepared in this study with variations of the native loop sequence CAR-[RDYRFDMGFD]-YWG and stabilizing strap motifs.