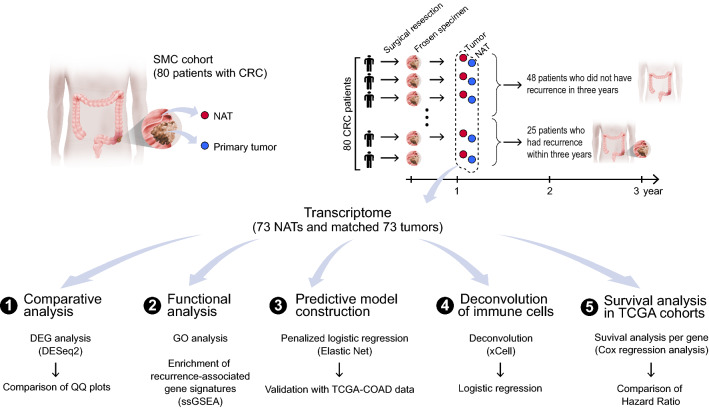
Fig. 1.
Study overview. RNA-seq data were produced from 160 surgical tumor and NAT samples from 80 Korean CRC patients. The total RNA-seq data, (i) tumor-derived transcriptomes and (ii) NAT-derived transcriptomes, were used to identify DEGs by comparing between RC and nonRC groups and functional analysis was done for the DEGs. The RNA-seq data of DEGs were used to construct recurrence prediction machine learning models. Subsequently, we investigated which machine learning prediction models constructed with NAT-based datasets or tumor-based datasets were superior for differentiating the recurrence states of patients with CRC. The two types of prediction models were then validated using the tumor-derived transcriptome data of 450 TCGA-COAD samples. Inferred immune cell composition were also used to compare which type of samples has more significant association with recurrence states with respect to infiltrated immune cell compositions. Finally, association of each gene with the survival of patient in different cancer types of TCGA cohorts was compared between NATs and tumor tissues