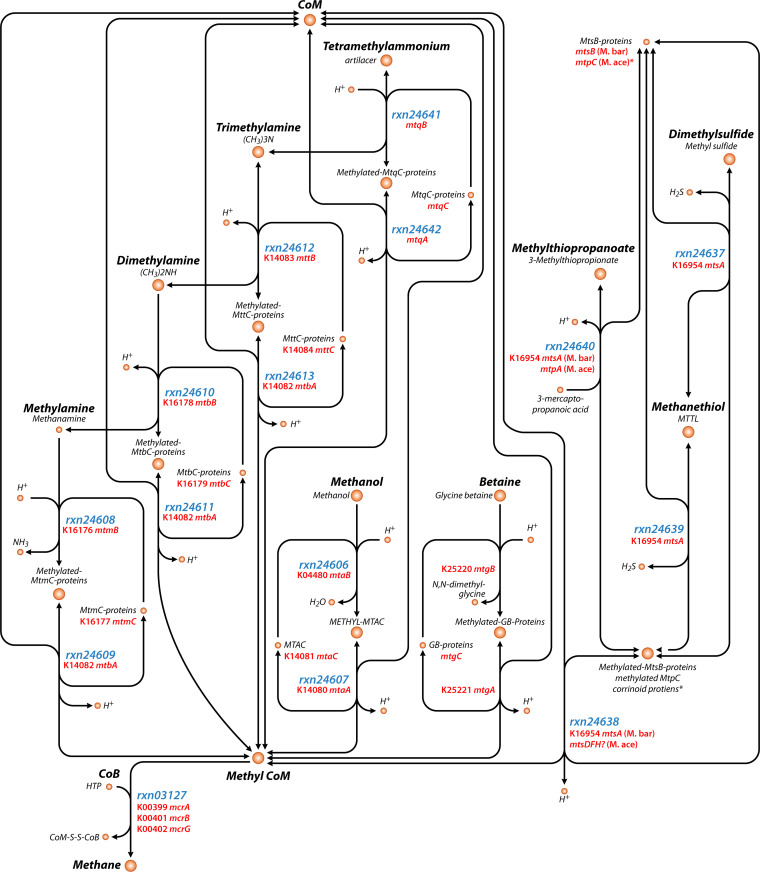
FIG 1.
Simplified map of methyl dismutation methanogenesis pathways showing ModelSeed compound names and reaction IDs as well as KEGG Orthology (KO) identifiers (when available) for the genes involved in the reactions. This map focuses on the methyl dismutation steps and does not show all of the proteins and pathways for energy conservation. The map was made with Escher based on MetaCyc, KEGG, and ModelSeed annotations and only includes those pathways in at least one of those databases as of the time of writing. Note that some gene names are described but do not yet have KO assignments. Also note that some reactions are not in ModelSeed (e.g., for betaine). The asterisk (*) indicates that methanogenesis from methylthiopropanoate differs in Ms. barkeri (“M. bar”) and Ms. acetivorans (“M. ace”), with Ms. acetivorans using MtsC proteins instead of MtsB proteins. A question mark (?) denotes hypothesized genes in need of further confirmation.