TABLE 1.
Methyl-based methanogenic substrates
| Namea | Abbreviation | Formula | Structureb | CH3-Dism. | CH3-Red. | Environmentsc | Genesd |
|---|---|---|---|---|---|---|---|
| Tetramethylammonium | QMA | (CH3)4N |
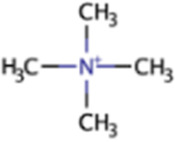
|
X | Marine sediment, industrial wastewater | mtqBCe, mtqAe | |
| Trimethylamine | TMA | (CH3)3N |
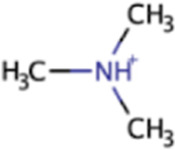
|
X | X | Marine sediment, hypersaline sediment, gut | mttBC, mtbA |
| Dimethylamine | DMA | (CH3)2NH |
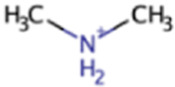
|
X | X | Marine sediment, hypersaline sediment, gut | mtbBC, mtbA |
| Monomethylamine | MMA | CH3NH2 |

|
X | X | Marine sediment, hypersaline sediment, gut | mtmBC, mtbA |
| Methanol | MeOH | CH3OH |

|
X | X | Marine sediment, freshwater sediment | mtaBC, mtaA |
| Glycine betaine | GB | C5H11NO2 |
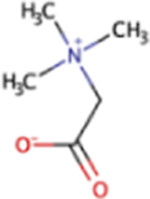
|
X | Marine sediment, hypersaline sediment | mtgBC, mtgA | |
| Dimethyl sulfide | DMS | (CH3)2S |
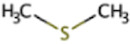
|
X | Marine sediment, hypersaline sediment | mtsAB, mtpC, mtsDEF | |
| Methanethiol | MT | CH3SH |

|
X | Marine sediment, hypersaline sediment, freshwater sediment | mtsAB, mtpC, mtsF | |
| Methylthiopropanoate | MMPA | C5H10O2S |
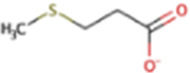
|
X | Marine sediment, hypersaline sediment | mtpP, mtsA, mtpA, mtpCAP |
Substrate names, abbreviations, formulas, structures, use in methyl dismutation (CH3-Dism.), use in methyl reduction (CH3-Red.), environments in which the substrates are present, and genes involved in the demethylation and methyl transfer to coenzyme M are shown.
Structure diagrams from ModelSEED.
Marine sediments here include ocean sediments as well as coastal and estuarine sediments, which are influenced by seawater.
Pathway-specific genes. All pathways would additionally need mcrABG. Some pathways would additionally require the Wood-Ljungdahl methyl branch and hydrogenases and membrane-bound proteins (Fig. 2).
Hypothesized but only demonstrated by one study.
