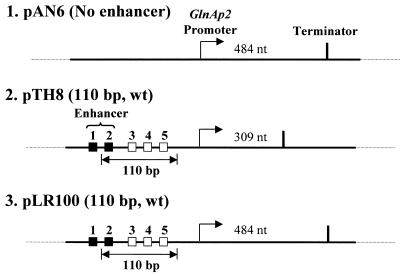
Figure 1.
Templates for analysis of the mechanism of glnAp2 promoter activation by NtrC-dependent enhancer. Plasmid templates having different enhancer–promoter spacing (constructs 2 and 3) and having no enhancer (construct 1). Strong and weak NtrC-binding sites are indicated by closed and open squares, respectively. Under our experimental conditions only the strong sites are occupied and contribute to the enhancer activity (10). The pTH8 and pLR100 plasmids (3.6 and 3.3 kb in size, respectively) have 110 bp wild-type (wt) enhancer–promoter spacing. The transcripts were terminated at the T7 terminator positioned over different distances downstream of the promoter. The lengths of the transcripts are indicated.