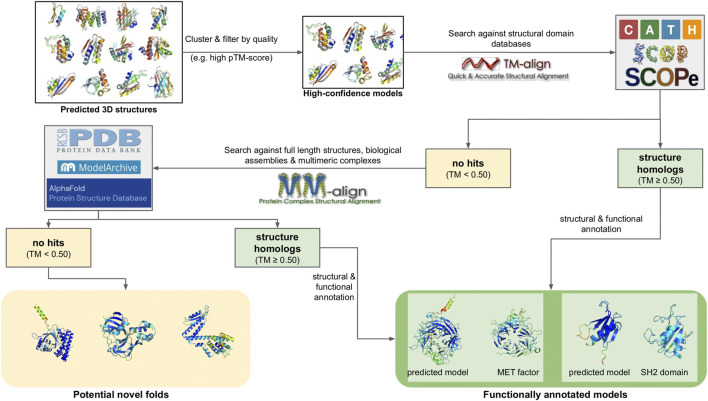
FIGURE 6.
Example structure search and functional annotation for a set of predicted 3D structures. In the first step, the models are filtered to keep only high-quality models, typically represented by a high predicted TM-score (pTM) value. The models are also clustered based on their structural similarity. The high quality, non-redundant set of models can then be searched against databases of structural domains (e.g., CATH-Gene3D, SCOP and SCOPe) with a fast, TM-score based method such as TM-align. Models with significant hits (TM-score ≥0.50) are functionally annotated based on their structural homologs. Models with no hits (TM-score <0.50) are further searched against databases of full-length structures (containing one or multiple domains), biological assemblies or protein-protein complexes (PDB, ModelArchive, AlphaFoldDB, etc.) with a multimeric complex-enabled search method such as MM-align. Again, models with significant hits are functionally annotated based on their homologs. Finally, models with no hits to any structural database can be considered as potential novel folds.