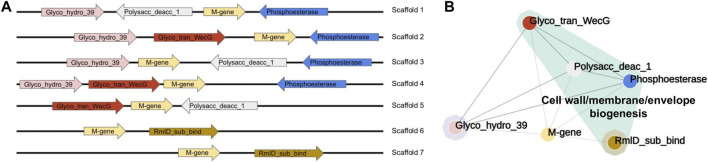
FIGURE 7.
Example of a gene neighborhood analysis for a cluster of unannotated metagenome sequences, represented as “M-gene”. (A) Simplified visualization of a synteny analysis for seven metagenome scaffolds, containing members of the M-gene cluster. Each gene is represented by an arrow and colored differently. The direction of the arrows represents the directionality of the ORFs in each scaffold. In the analyzed scaffolds, the M-gene ORF co-occurs with a number of other protein-coding genes, each corresponding to a Pfam domain. (B) Gene co-occurrence network, based on the results of the synteny analysis. Each node represents a protein-coding gene and is colored using the same scheme as in (A). Edges (interactions) between nodes are derived based on the co-occurrence of their genes in the same scaffold. As it can be seen, the unannotated metagenomic cluster (M-gene) co-occurs with a tightly connected group of Pfam domains (Phosphoesterase, Glyco_tran_WecG, Polysacc_deacc_1 and Glyco_hydro_39), which are all found in the same scaffolds alongside M-gene members. In addition, M-gene co-occurs with RmID_sub_bind. Notably, four of the co-occurring protein domains are in the same functional category (Cell wall/membrane/envelope biogenesis), as indicated by their annotation in COG. This could mean that the unannotated M-gene cluster may participate in this function as well. The network was constructed using NORMA (Karatzas et al., 2022b).