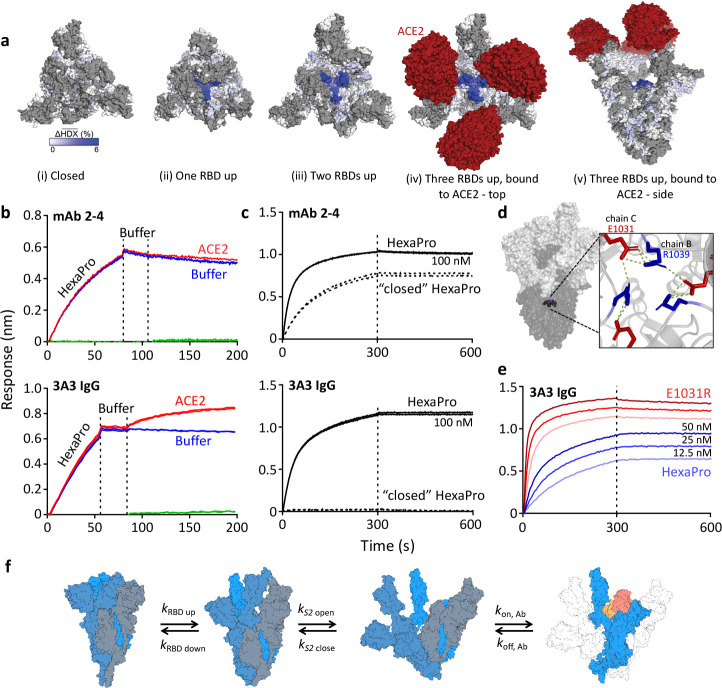
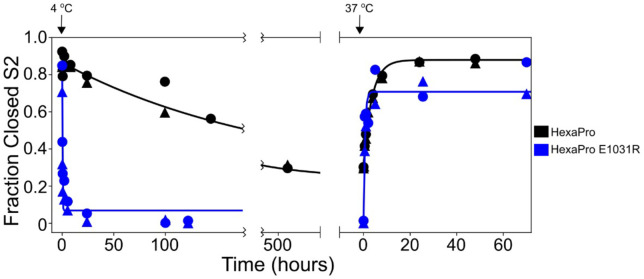
Figure 2. The hinge epitope is accessible only in an RBD-up and S2-open spike conformation.
(a) Trimeric SARS-2 spike in various conformations colored according to difference in deuterium fractional uptake between SARS-2 HexaPro spike alone and with 3A3 IgG. The hinge epitope within S2 is colored dark blue in structures of wild-type SARS-2 spike in the (i) three RBDs down or closed conformation (PDB: 6XR8) and in structures of stabilized spike with (ii) one RBD up (PDB: 6VSB), (iii) two RBDs up (PDB: 7A93), or (iv) three RBDs up while bound to ACE2 (red) in top-view and (v) sideview (PDB: 7A98). Residues lacking coverage in the HDX experiment are indicated in gray. (b) Antibody 3A3 (bottom) or control mAb 2–4 (top) were coupled to anti-Fab BLI sensors and allowed to capture HexaPro or nothing (buffer, green line), then dipped into buffer (baseline), and finally dipped into ACE2-Fc (ACE2, red) or nothing (buffer, blue). (c) BLI binding of immobilized control mAb 2–4 (top) or antibody 3A3 (bottom) to 100 nM HexaPro (solid) or HexaPro locked into the ‘closed’ conformation (dashed). Vertical dashed lines indicate start of dissociation phase. (d) The network of hydrogen bonds formed by residues E1031 and R1039 across protomers deep in the S2 core is shown on intact HexaPro spike and in detail in a top view (PDB: 6XKL). (e) Antibody 3A3 was coupled to anti-Fc BLI sensors and allowed to bind HexaPro or E1031R HexaPro (E1031R) spike protein. All BLI data are representative of biological duplicates. Each experiment was repeated in technical duplicate except e, which was tested once at each concentration to allow all data to be collected simultaneously for direct comparison. (f) Model of the kinetic changes required for antibody binding to the hinge epitope, including conversion of the RBDs into the up position and some degree of opening of the S2 domain in addition to typical antibody association and dissociation kinetics (generated using PDB 6XV8 and 7A98).