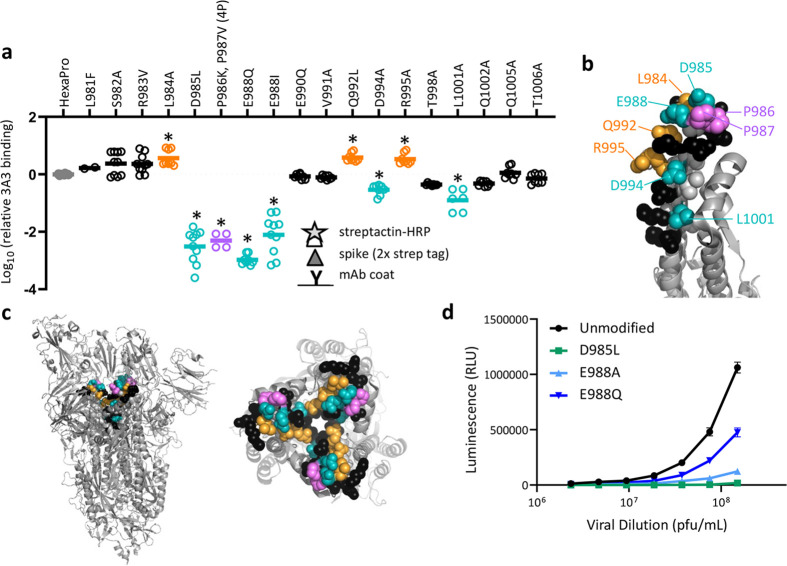
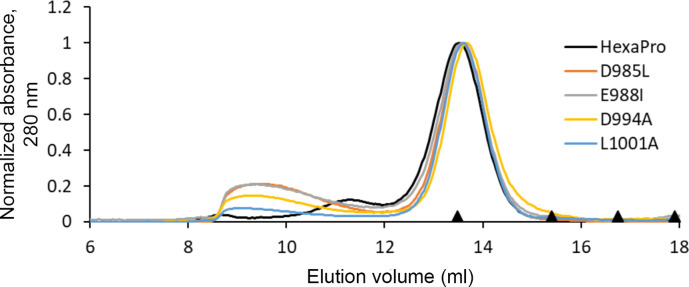
Figure 3. SARS-2 spike residues 985–988 are recognized by 3A3 and impair spike function upon substitution.
(a) Residues important for 3A3 binding were identified by single residue changes in HexaPro that increased or decreased binding to 3A3 relative to HexaPro. Each variant was tested with duplicate technical replicates in 2–6 independent ELISA assays. Significance relative to unaltered HexaPro was determined by ANOVA with post-hoc Tukey–Kramer test with α = 0.01; data meeting this criterion indicated by *. (b) Location of the residue changes altering binding to 3A3 shown in the HexaPro spike (6XKL) monomer and in (c) intact spike (side view) and the S2 domain (top-down view). All epitope residues (980–1006) are shown in space-fill, with residues colored according to their effect on 3A3 binding: improved binding (orange), reduced binding (teal), no effect (black), and those not altered (gray). The 2P stabilizing mutations within the hinge epitope are displayed in purple. (d) The infectivity of lentivirus pseudotyped with unmodified D614G SARS-2 spike or variants with D985L (green), E988A (light blue), and E988Q (dark blue) substitutions was compared by luciferase activity. Data shown are the mean luminescence with standard deviation of three technical replicates.