Fig. 1 |. Efficient capture of gRNAs for CaRPool-seq.
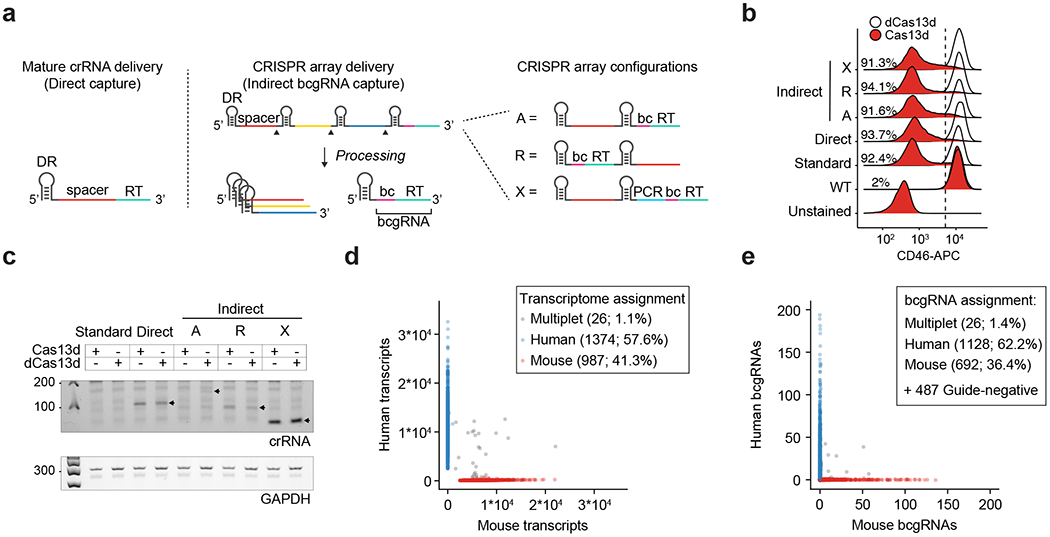
a, Scheme of direct and indirect array-based gRNA capture approaches. Direct capture uses a reverse transcription (RT) handle added directly downstream to the gRNA. For the indirect capture method, a bcgRNA is captured as a cleavable part of a CRISPR array. Three different CRISPR array configurations (A, R and X) have been tested (bc, barcode; PCR, PCR primer annealing site; A, array; R, reversed array configuration; X, extra PCR handle). b, Density plots showing the CD46-APC signal on Cas13d-mediated CD46 knockdown (red) and dCas13d-mediated controls (white) using the four CaRPool-seq configurations described in a, as well as standard gRNA. The CS1 reverse transcription handle was used in all cases. N > 5,000 cells examined per sample. c, PCR amplicons of reverse-transcribed crRNAs from lentivirally infected cells used in b of one representative experiment. Expected product sizes: Direct capture 109 bp, A-type and R-type array 99 bp, X-type array 52 bp, unprocessed A-type array (159 bp). d, Species-mixing experiment profiling 2,387 HEK293FT-Cas13d or mouse NIH/3T3-Cas13d cells lentivirally transduced with CRISPR array virus. The CRISPR array includes a NT gRNA and a bcgRNA in X-type configuration. Axes show the number of transcripts associated with each cell barcode. Datapoint colors and boxed labels are assigned based on transcriptome classification (Methods). e, Number of bcgRNAs associated with each cell barcode. Datapoint colors are based on transcriptome classification, and boxed labels are based on observed bcgRNA.
