Abstract
The documentation of the change in the number and appearance of pigmented cutaneous lesions over time is critical to the early detection of skin cancers and may provide preliminary signals of efficacy in early-phase therapeutic prevention trials for melanoma. Despite substantial progress in computer-aided diagnosis of melanoma, automated methods to assess the evolution of lesions are relatively undeveloped. This report describes the development and narrow validation of mathematical algorithms to register nevi between sequential digital photographs of large areas of skin and to align images for improved detection and quantification of changes. Serial posterior truncal photographs from a pre-existing database were processed and analyzed by the software, and the results were evaluated by a panel of clinicians using a separate Extensible Markup Language‒based application. The software had a high sensitivity for the detection of cutaneous lesions as small as 2 mm. The software registered lesions accurately, with occasional errors at the edges of the images. In one pilot study with 17 patients, the use of the software enabled clinicians to identify new and/or enlarged lesions in 3‒11 additional patients versus the unregistered images. Automated quantification of size change performed similarly to that of human raters. These results support the further development and broader validation of this technique.
Abbreviation: CI, confidence interval
Introduction
Cutaneous melanoma may arise either de novo or from nonobligate precursor lesions such as atypical/dysplastic nevi (Shain and Bastian, 2016). The assessment of changes in the number and morphology of pigmented cutaneous lesions over time is used in many centers to improve the early detection of melanoma (Banky et al., 2005; Mar et al., 2017). In addition, we and others have hypothesized that changes in the appearance of nevi may provide preliminary signals of efficacy in early-phase therapeutic prevention (chemoprevention/immunoprevention) trials for melanoma (Maguire and Kirkwood, 2020), analogous to how changes in the number of actinic keratoses provided a preliminary signal of the efficacy of nicotinamide in the prevention of nonmelanoma skin cancer (Chen et al., 2015; Surjana et al., 2012). An objective, reproducible means of measuring changes in the appearance of cutaneous pigmented lesions will facilitate both clinical and research applications.
The assessment of the change of lesions in serial digital images by human observers is complicated by numerous factors (Schneider et al., 2019). Factors relating to the images themselves may include differences in scale, body habitus, posture, and lighting between the serial photographs. Factors relating to the human evaluator include differences in provider experience as well as time pressures posed by clinical workload and competing responsibilities. These issues contribute to the limitations of human perception of subtle changes in multiple lesions simultaneously under suboptimal conditions. This is particularly relevant for nevi, a subset that is dynamic, but the majority is stable or shows minor or no change over time when assessed by human reviewers (Banky et al., 2005). We propose that a computer vision approach can help to mitigate these problems and produce a more accurate assessment of change in cutaneous lesions.
Despite the use of total body digital photography for at least 20 years to document the presence of cutaneous lesions (Hornung et al., 2021) as well as substantial recent progress in computer-aided diagnosis of lesions in clinical images (Dick et al., 2019; Haenssle et al., 2020; Tschandl et al., 2019), automated methods to characterize the evolution of skin lesions are still lacking. Automated image registration refers to the use of computational models to correlate features between two or more images. In many cases, the process also includes transforming one or both of the images so that the features can be aligned for improved comparison. Numerous techniques for automated image registration have been developed over the past few decades and are of great interest in a variety of fields, including remote sensing (e.g., detection of changes by orbiting satellites) (Tondewad and Dale, 2020) and medical imaging (Guan et al., 2018). Registration and alignment of photographs of human skin pose a more challenging problem than registration in other domains because skin requires simultaneous accommodation of changes in weight, wrinkling, hair, scale, skin tone changes, lighting, translation, rotation, and warping.
The purpose of this study was to develop and preliminarily validate a computer vision approach to facilitate the detection and quantification of changes in nevi in serial digital photographs.
Results
Features of the pilot study
Image pairs of the posterior trunk from 24 patients were obtained from the archival image database, of whom seven were excluded from analysis by DermaViz because they did not meet the criteria for inclusion listed in Materials and Methods. The most common reasons for exclusion were image pairs with too few features to register; images with poor acquisition, such as uneven/poor cropping; and durations >6 years between images.
Of the patients whose photographs were analyzed, the mean age at the time of the first photograph was 38.4 years (range = 8.8‒79.0 years; SD = 16.8 years). Seven patients were men, and 10 were women. Recorded Fitzpatrick skin types were Fitzpatrick 1 (four patients), Fitzpatrick 1‒2 (two patients), Fitzpatrick 2 (eight patients), Fitzpatrick 2‒3 (one patient), and unknown (two patients). Among patients for whom historical information was available, 14 carried a previous diagnosis of melanoma. The remaining three patients did not have a previous histopathologic diagnosis available. One patient had two pairs of photographs, so there were 18 total image pairs included in the analysis. The mean time between paired photographs was 3.4 years (range = 0.5‒7.2 years; SD = 1.4 years).
Change assessment in posterior truncal images
To assess whether registration and alignment by the software improved clinicians’ ability to detect a change, three practicing dermatologists and two medical oncologists involved in the care of patients with melanoma reviewed the paired serial full-back images and enumerated the number of pigmented lesion pairs in several categories: decreased, disappeared, surgically removed, increased, or appeared. To reduce the potential for bias due to carry-over effects, the images in each set (unregistered and registered) were presented in random order, and each set of images was presented during different viewing sessions, with the unregistered images shown first. The mean delay between viewing the first and second sets of images was 14 days (range = 0.167–40 days; SD = 17 days).
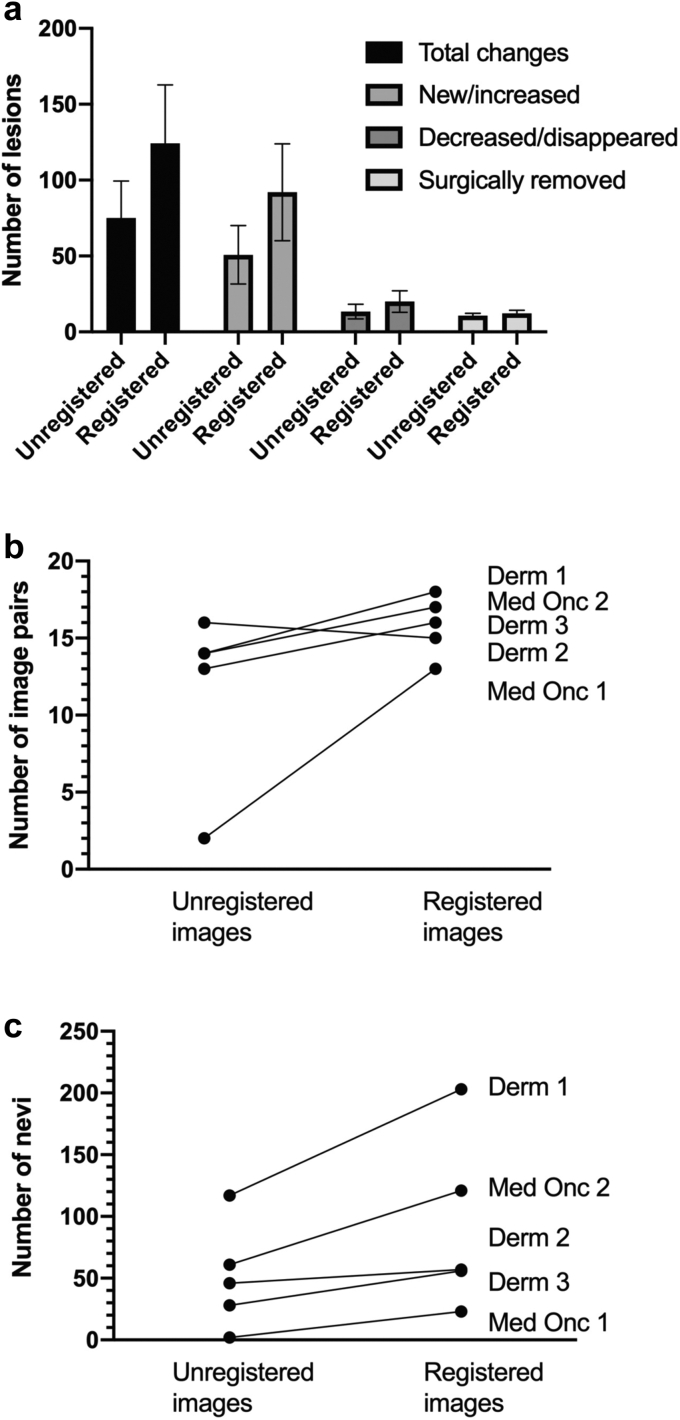
Clinician use of DermaViz increased the number of lesions detected in several categories: total changed lesions, new/increased lesions, decreased/disappeared lesions, surgically removed lesions (Figure 1a), and image pairs with new/increased lesions detected (Figure 1b). The increase in the detection of total changed lesions (average = 1.7-fold increase) was primarily driven by a 1.8-fold average increase in the detection of new or increased lesions (Table 1). The changes in decreased/disappearing lesions and surgically removed lesions were less consistent and of lower magnitude.
Figure 1.
Effects of DermaViz on detection of change by clinicians. (a) Nevus change detection by category. (b) New/increased nevi detected. (c) Image pairs with new/increasing nevi detected. Error bars represent SEM. Derm, dermatologist; Med Onc, medical oncologist.
Table 1.
Numerical Results of Change Assessment Exercise
| Provider | Image Pair Type | Time between Viewing Registered and Unregistered Images (d) | Number of Total Changes Detected | Number of New/Increased Lesions Detected | Number of Decreased/Disappeared Lesions Detected | Number of Surgically Removed Lesions Detected | Number of Image Pairs with New/Increased Lesions Detected |
|---|---|---|---|---|---|---|---|
| Dermatologist 1 | Unregistered | 21 | 158 | 117 | 30 | 11 | 14 |
| Registered | 244 | 203 | 25 | 16 | 18 | ||
| Dermatologist 2 | Unregistered | 10 | 75 | 46 | 17 | 12 | 16 |
| Registered | 82 | 57 | 11 | 14 | 15 | ||
| Dermatologist 3 | Unregistered | 40 | 48 | 28 | 6 | 14 | 13 |
| Registered | 75 | 56 | 10 | 9 | 16 | ||
| Medical oncologist 1 | Unregistered | 1 | 10 | 2 | 3 | 5 | 2 |
| Registered | 37 | 23 | 8 | 6 | 13 | ||
| Medical oncologist 2 | Unregistered | 0.167 | 84 | 61 | 11 | 12 | 14 |
| Registered | 183 | 121 | 46 | 16 | 17 | ||
| Total mean | Unregistered | 14.4 | 75.0 | 50.8 | 13.4 | 10.8 | 11.8 |
| Registered | 124.2 | 92.0 | 20.0 | 12.2 | 15.8 | ||
| Total SD | Unregistered | 16.6 | 54.6 | 43.0 | 10.7 | 3.4 | 5.6 |
| Registered | 86.0 | 71.5 | 16.0 | 4.5 | 1.9 | ||
| Fold difference between registered and unregistered | 1.7 | 1.8 | 1.5 | 1.1 | 1.3 | ||
DermaViz improved the detection of new or increased lesions by all providers (Figure 1c), despite differences in the specialty of the provider, the amount of time elapsed between viewing the unregistered and registered images, and the baseline number of images detected in each category. Complete numerical results are provided in Table 1.
Registration produced statistically significant improvements in the identification of new and increased lesions (P < 0.001) and in the identification of image pairs with changes detected (P < 0.001) versus unregistered images, as assessed by linear mixed effects models. The ORs for changes detected between registered and unregistered images was 4.85 (95% confidence interval [CI] = 2.06‒11.39).
Assessment of sensitivity and characterization of detected lesions
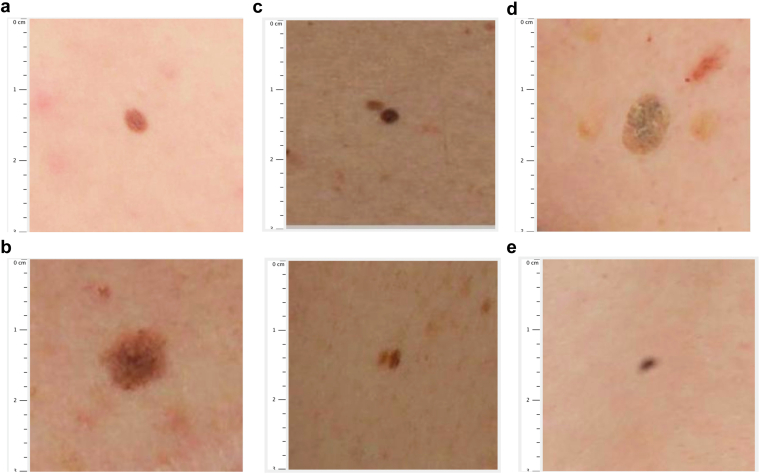
To evaluate the accuracy of DermaViz software to identify nevi and other cutaneous lesions, we compared the results of lesion identification by the software with those of clinician reviews of 46 randomly selected subfields of the posterior truncal images by two medical oncologists and one advanced practice provider, all of whom were involved in the care of patients with melanoma as detailed in the methods. A consensus of three of the three clinicians was considered a gold standard of lesion identification in this study. Of note, there were no disagreements between clinicians regarding nevus identification. Figure 2a and b shows the representative images of common and atypical nevi.
Figure 2.
Spectrum of lesions detected by DermaViz. (a) Common melanocytic nevus. (b) Atypical melanocytic nevus. (c) Adjacent nevi counted as a single nevus. (d) Nonmelanocytic lesion (seborrheic keratosis). (e) Indeterminate lesion (likely artifact such as dust on lens). Images are screenshots from the clinician-facing portion of the Validator application, taken by a clinical reviewer.
For lesions over 2 mm where three of the three clinicians identified a nevus, DermaViz agreed for 67 of 69 lesions. This corresponds to a false negative rate of 2.90% (exact 95% CI = 0.8‒7.5%) and a sensitivity of 97.1% (exact 95% CI = 92.5‒9.2%). The two nevi missed by DermaViz were both small nevi that were counted together with immediately adjacent larger nevi (Figure 2c).
As noted earlier, the current version of DermaViz is not intended to classify lesions by type; however, this information can be added by clinicians using the Validator application. Of the 72 lesions over 2 mm identified by DermaViz in the nevus centroid exercise, 67 (93%) were identified as melanocytic nevi by three of the three clinicians. In the nevus size exercise, in which the lesions were selected through different criteria as noted in Materials and Methods, a lower percentage of lesions were found to be melanocytic (average of 66 of 97 lesions or 68%).
Non-nevus lesions detected by DermaViz were frequently classified by clinicians as seborrheic keratoses (examples are shown in Figure 2d), but other nonmelanocytic structures were identified by the software as well, including occasional suspected artifacts including possible dust on the camera lens (Figure 2e).
Assessment of the accuracy of registration
Two methods were used to assess whether the DermaViz software had successfully registered nevi between time points for individual patients. The first method involved a change detection exercise involving aligned posterior truncal photographs displayed as sequential PowerPoint slides. The five clinician reviewers who participated in this exercise determined that on average, 17 of 18 images (range = 15‒18) had greater than 90% of lesions correctly aligned. The second method involved comparison in the Validator application of 97 paired fields, which included a variety of lesion types, including those with increasing, decreasing, appearing, and disappearing lesions. Of the 97 paired fields, 87 clearly had lesions identified in both images, whereas the remaining 10 had lesions identified that either appeared or disappeared. The three clinician reviewers who completed this exercise determined that all 87 images included in this analysis that had paired lesions in the subsequent image had been correctly registered. Of note, lesions on the edges of the images had been specifically excluded from this analysis.
Despite most lesions being correctly registered, there were still occasional errors. Supplementary Movie S1 illustrates a typical image pair where most lesions are correctly registered, but there are a limited number of nevi on the edges of the image that are incorrectly registered and not properly aligned.
Assessment of the accuracy of size change
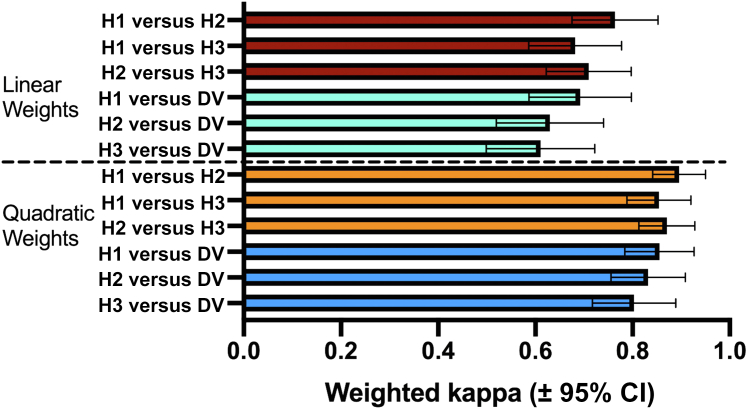
To compare the assessment of size change by humans with the DermaViz quantification, three clinicians (two medical oncologists and one advanced practice provider) were shown sequential zoomed-in views of lesion pairs in the Validator interface, which were selected as described in the Materials and Methods section. Clinicians were asked to characterize changes in the size of the lesions on an ordinal scale. Inter-rater reliability between a clinician and the other and between each clinician and DermaViz was computed using Cohen’s kappa. Data are presented as weighted kappas. We feel that quadratic weighting relates more to the current analysis because it gives harsher penalties to larger disagreements and also because it produces results similar to those of Fleiss intraclass correlation, another commonly used method of inter-rater reliability (Fleiss and Cohen, 1973). However, we have reported both linear and quadratic weights because of varying opinions in the literature, over which weighting system is preferable (Vanbelle, 2016).
As shown in Figure 3, clinicians showed moderate (with linear weights) to good (with quadratic weights) agreement with each other. DermaViz also showed moderate (with linear weights) to good (with quadratic weights) agreement with the clinicians. The weighted kappas were slightly lower for DermaViz than for the human raters; however, the 95% CIs overlapped such that the difference was not significant for any rater pair. The numerical kappa values between raters as well as the individual 95% CIs are shown in Tables 2 and 3.
Figure 3.
Size change assessment by DV compared with that of human observers. Bars represent weighted kappas between all human observers (H1, H2, and H3) and between each human observer and DV. CI, confidence interval; DV, DermaViz; H, human.
Table 2.
Linear Weighted Cohen’s Kappa between Each Pair of Two Raters for Size Change Exercise, ±95% Confidence Intervals
| Rater | H1 | H2 | H3 | DV |
|---|---|---|---|---|
| H1 | — | 0.7636 (0.651‒0.8522) | 0.6819 (0.5862‒0.7777) | 0.6924 (0.5870‒0.7978 |
| H2 | — | — | 0.7095 (0.6221‒0.7970) | 0.63 (0.5195‒0.7404) |
| H3 | — | — | — | 0.6106 (0.4991‒0.7221) |
| DV | — | — | — | — |
Abbreviations: DV, DermaViz; H, human.
Table 3.
Quadratic Weighted Cohen’s Kappa between Each Pair of Two Raters for Size Change Exercise, ±95% Confidence Intervals
| Rater | H1 | H2 | H3 | DV |
|---|---|---|---|---|
| H1 | — | 0.8958 (0.8415‒0.9500) | 0.8542 (0.7882‒0.9202) | 0.8553 (0.7840‒0.9267) |
| H2 | — | — | 0.8708 (0.8131‒0.9284) | 0.8322 (0.7558‒0.9087) |
| H3 | — | — | — | 0.8030 (0.7172‒0.8888) |
| DV | — | — | — | — |
Abbreviations: DV, DermaViz; H, human.
Discussion
In this study, DermaViz automated image registration facilitated the detection and quantification of changes in size and number of pigmented lesions of interest in sequential digital photographs by melanoma clinicians. The software helped clinicians identify numerous changes that were missed in the original unregistered images. Importantly, because the DermaViz approach uses photographs of large areas of skin, it potentially offers an important advantage over the current clinical practice of change assessment through serial dermoscopy of single-pigmented lesions (Morris et al., 2017). Pigmented lesions of concern must be prespecified by physicians before they can be evaluated through dermoscopy; such focused assessment will miss changes in lesions that were not specifically selected for baseline imaging (Fuller et al., 2007). DermaViz inherently tracks multiple pigmented lesions contained in the photographs and computes an accurate relative change between sequential photograph dates. A promising combination strategy might be an initial step of automated image analysis of digital camera photographs, followed by a focused dermoscopy of concerning lesions.
There have been relatively few previous attempts at registering skin lesions in digital photographs of large areas of the skin, and our experience with DermaViz compares favorably with these attempts. An early approach for registration represented pigmented lesions as points on a lesion map, identified initial matching lesion pairs in the sequential images, selected four matching image pairs near the image corners using an iterative method designed to reduce mismatches, and then transformed one image to match the lesion map of the other image using the four selected points (Mcgregor, 1998). Results were promising; yet, the testing used data from the same date, and although it accounted for some posture changes, it did little to handle lighting or scale variations. Another approach to registration included alignment using structured geometric and tensor-based models; yet, this approach was not shown to be robust to changes in posture or scale (Mirzaalian et al., 2016). In the Canfield approach (Korotkov et al., 2015), the lesion registration and flickering are robust, yet this was shown only with specific nevus pairs as opposed to detecting a change in larger body regions, such as the posterior trunk, leg, or arm. Canfield and other approaches have used three-dimensional total body photography (Hornung et al., 2021), which minimizes variability in lighting and posture because of constraints on the patient. The Fotofinder technology includes advanced image acquisition hardware and lesion classification (Del Rosario et al., 2018; Levy et al., 2003); however, we found no publication noting that this technology performs automated registration and detection of change. For both the Canfield and Fotofinder approaches, the use of expensive, specialized imaging hardware may limit their use to specific clinical settings. The approach used by DermaViz is one that may be affordably implemented by a variety of dermatology and potentially even primary care settings. Furthermore, DermaViz could theoretically be used to retrospectively compare archival digital camera images, whereas approaches requiring advanced image acquisition devices are limited by the relatively recent introduction of these devices.
The DermaViz software can theoretically be used to improve the comparison of images obtained with mobile devices such as smartphones and tablets, whose camera capabilities have improved markedly in recent years. Supplementary Movie S2 shows the successful registration and alignment of images obtained by an iPhone 11. There has been considerable interest in the use of smartphone applications to facilitate computer-aided diagnosis and monitoring of suspicious skin lesions, although the results generated by the software have so far not been shown to be of clinical utility (Freeman et al., 2020; Matin and Dinnes, 2021). A recent study showed that the parameters of commercial digital cameras and smartphones could be modified to increase the accuracy of the images, generally by disabling some automatic settings that were intended to produce more visually appealing images for consumers but can reduce accuracy (Dugonik et al., 2020). For optimal results with DermaViz analysis of smartphone images, it will be important to specify acquisition protocols to promote accuracy and consistency between images acquired over time. This process may be aided by further progress in developing standards for the imaging of skin lesions (Finnane et al., 2017). Given the anticipated need for at least some standardization in image acquisition, we anticipate that smartphone images will need to be taken in clinical settings with appropriate training, and we do not anticipate that patient-obtained smartphone photographs will generally be suitable for use with DermaViz. The main implication of using DermaViz with smartphone photographs is that it would greatly expand the number and type of physician offices that could use the software.
Several images in this study could not be accurately registered because of issues with the suboptimal quality and heterogeneity of the archival images; however, it should be noted that the images were not acquired for this purpose and did not have consistent acquisition parameters. We expect that prospective photography using even simple, uniform acquisition parameters will result in a higher percentage of images that can be registered by DermaViz. Although there is no requirement for a particular resolution of camera/image needed to use DermaViz, we expect that high resolution and in-focus images will provide additional detail that will increase the number of image pairs that can be successfully registered as well as the accuracy of the registration.
A notable feature of this study was the use of sequential images from patients that were taken as part of an image and nevus biobanking protocol at our institution. This differs from several other studies that used artificial means of generating changes in size and number of cutaneous lesions before analysis, either by drawing them on the skin (Guido et al., 2018) or by digitally modifying the original images to introduce the changes (Navarro et al., 2019). The use of artificial nevi and/or digital changes to nevi may not reflect a real-world situation and may introduce artifacts and/or bias owing to assumptions made when creating or modifying the lesions. The use of sequential patient images is more relevant to real clinical situations, but it also makes it more challenging to identify an appropriate gold standard and/or define a ground truth with which to compare the results from DermaViz. A common gold standard method that has been used to evaluate the quality of segmentation in unmodified clinical images is the Society for Imaging Informatics in Medicine-International Skin Imaging Collaboration Melanoma Classification Challenge (Rotemberg et al., 2021), which has been used to evaluate the segmentation and classification capabilities of numerous other image analysis tools. We did not use this for this study because our images are camera photographs of the entire posterior trunk, whereas the images in the Society for Imaging Informatics in Medicine-International Skin Imaging Collaboration are generally dermatoscopic images of single skin lesions. As previously noted, the DermaViz method enables tracking of any nevus that changes in the photograph, including those that were not identified as atypical on earlier dates. Furthermore, the assessment technique used by DermaViz is inherently different from techniques focused on the analysis of single lesions because the coregistration of multiple lesions contributes to a higher accuracy of change assessment than if DermaViz was used to analyze individual nevi. Therefore, the segmentation of individually chosen lesions is not fully indicative of the capabilities of DermaViz. It could be argued that the use of clinical observers, as was done in this study, is not an ideal gold standard either, particularly because the automated analysis can leverage additional features of the image such as altered color spectra that are not visible to human observers. The small discrepancies between human observers and DermaViz may reflect deficiencies in human perception or deficiencies in the imaging program, and it is currently difficult to separate these two possibilities. We plan to address this in further studies, for example, by presenting the human clinicians with modified images that represent the optimal color spectrum that DermaViz uses to detect change.
The conclusions of this study may have been affected by the limitations of the image dataset used and the DermaViz software itself, both of which we hope to address in the future. Regarding the limitations of the image dataset, these historical photographs were taken by research assistants. There were variations in background, lighting, focus, and exposure, and it is conceivable that changes in imaging parameters might contribute to some of the observed changes. The images did not consistently incorporate a measurement ruler and used no form of calibration for color or lighting. There was a wide range of follow-up intervals between images. Finally, the image dataset comprised a small number of patients at a single institution. To address these potential issues in future studies, we have designed and are now including a new measurement tape (including a ruler) and a color balance feature in every image for improved characterization of size, border, and color changes. At the UPMC Hillman Melanoma Program, photographs have for the past 2 years been taken by a single professional photographer with a high-resolution 12-bit digital camera and standardized lighting and formatting. Prospective studies will obtain images at consistent, prespecified time intervals and ideally include involvement with multiple clinical sites to improve external validity. It will be important to include more patients, including patients with diverse skin types. Regarding the DermaViz software itself, there are four primary limitations of the current version of DermaViz. First, the automated algorithms do not yet determine lesion type, although clinicians are readily able to input this information using the Validator interface. Future versions of DermaViz could include a computer-aided diagnosis feature, and/or the current software could be combined with one of many existing computer-aided diagnosis programs. Second, normalization of color and scale between images using the ruler/color balance tape has not yet been validated because we have only recently started using the updated tapes. Third, the technology has so far not been extensively used to characterize lesions on curved surfaces such as limbs, and this will need to be explored more in future studies. Finally, DermaViz’s metrics to quantify border irregularity, asymmetry, and color will need to be validated before their accuracy/clinical relevance can be shown. Of note regarding the last point, we expect that the current version of DermaViz, which primarily measures changes in size and the number of lesions, will still be useful for a variety of applications even before the additional quantitation of borders, color, and asymmetry has been validated.
The intended use of DermaViz at this point is in additional research studies in clinical office settings. These subsequent studies are needed to validate the software more broadly for both clinical and research uses. In terms of clinical use, a critical question that will eventually need to be addressed prospectively is whether the increased ability to detect changes noted in this study results in differences in patient care, such as different rates of biopsy and ultimately different rates of skin lesion diagnosis. This type of impact analysis could be achieved through a larger prospective study that compares the care of patients whose providers used DermaViz with that of a separate group of patients whose providers used traditional methods of monitoring skin lesions. Future studies should consider the time course of the changes because more rapid changes would generally be more concerning. It will be important to also assess the possible effects on overdiagnosis and physician fatigue, which are risks of this approach. In terms of research use, further validation of DermaViz will involve using the software to analyze changes in size and number (and later color, border, etc.) between patient groups that are prospectively exposed to different treatment conditions to show whether changes in nevus morphology can be detected between those groups. After discussion with numerous melanoma experts, we have hypothesized that morphologic changes in size and number of atypical/dysplastic nevi would be the most likely useful surrogate for the efficacy of therapeutic prevention agents (chemoprevention/immunoprevention) (Maguire and Kirkwood, 2020), although it remains to be shown prospectively whether any changes observed correlate with other biomarkers of melanoma development or with meaningful clinical outcomes. To this end, we intend to use DermaViz in a planned phase II study of 1 year of treatment with the broccoli-derived agent sulforaphane versus placebo in patients with multiple atypical nevi and a history of melanoma (EA6201).
This publication of the description and narrow validation of DermaViz software may raise awareness of the technology in the dermatology and oncology communities, which we hope will facilitate the prospective studies that will ultimately be needed to more fully develop and validate this technology.
Materials and Methods
Please note that we have endeavored to conform to a published consensus checklist for evaluation of image-based artificial intelligence reports in dermatology (Daneshjou et al., 2022) to the extent that is possible given the proprietary nature of the DermaViz software.
Description of DermaViz
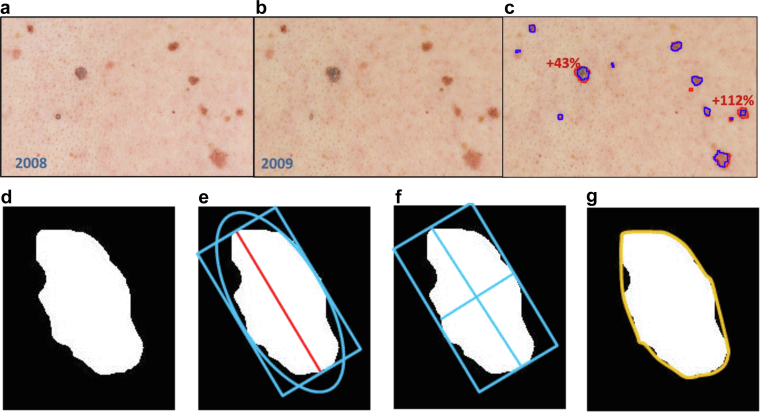
The registration approach used by the DermaViz software is composed of four main steps, for which the skin lesions are considered points in a coordinated system defined as the skin space. First, the technique computes a coarse estimate of the scale and rotation of the points on the basis of the logarithm of potentially matched point differences. Second, it computes a low-resolution translation estimate, leading to point matches and nonlinear mapping. Third, the algorithm computes a high-resolution translation estimate over subimage tiles of the entire image, leading to improved point matches and nonlinear mapping. Finally, the algorithm performs cross-checking of the point matches to eliminate false matches, followed by a reiteration of these steps until all of the point matches are confirmed as accurately matched. Overall, if corresponding paired images have at least 50% common overlap in the recorded skin surface area, then the algorithms can accommodate resolutions ratios as disparate as 1:3 linear, 1:9 area, and angular rotations of ±10 degrees along with mild key stoning and additional translation shifts. Figure 4 shows registered and aligned areas of the posterior trunk of a patient photographed 1 year apart (Figure 4a and b) as well as segmentation boundaries and measured changes (Figure 4c). The aligned images can be toggled back and forth within the same frame to better visualize changes in individual lesions. Supplementary Movies S1 and S3 show representative clinical images of two patients before and after processing with DermaViz, displayed in the toggle view. Supplementary Movie S2 presents similar before-and-after processing image pairs acquired with a mobile phone device.
Figure 4.
Demonstration of DermaViz software. (a) Initial timepoint. (b) Subsequent time point after registration and alignment. (c) Overlaid segmentation boundaries and example change quantification. (d) Delineation of border. (e) Delineation of diameter. (f) Delineation of major and minor axes. (g) Delineation of convex hull. Figure 4d‒g is reproduced from Tahata et al. (2018). Images were provided by Veytel.
The segmentation approach to distinguish skin lesions from the background skin is an algorithm that converts the skin’s red‒green‒blue image into three separate images for hue, saturation, and intensity (Blotta et al., 2011). Nevus filtering-thresholding algorithms for each of the hue, saturation, and intensity domains are applied individually, with filters tuned for the statistics of the nevus and background skin regions, respectively. The resulting three images are recombined to create a unified segmentation of the nevus/skin boundary.
Once lesion segmentation is defined, size features are readily calculated for the area, diameter, and perimeter. The basis of size and other measurements is illustrated in Figure 4d‒g (reproduced from Tahata et al. [2018]), with the nevus shown in white and a rectangle enclosing the nevus that has the same major axis as the ellipse. The area is computed in square millimeters using the pixels within the border. The diameter is the extent of the lesion in the direction of the major axis of an ellipse that has the same second moments as the nevus, measured in millimeters (mm). The perimeter is the measure of the distance around the border in millimeters. Table 4 details the classic ABCDE features of melanoma clinical presentation (Glazer et al., 2017) as well as a general description of how they are quantified by the DermaViz software. Of these, the size-related measurements are the main focus of this study. It should be noted that in its current version, DermaViz does not classify lesions by type.
Table 4.
ABCDE Features and Measurement by DermaViz
| Quality | Type | Description |
|---|---|---|
| A Asymmetry |
Component | A measure of the asymmetrical components of the quadrants of the nevi. |
| B Border irregularity |
Variation | A measure of the difference in the ratio of the nevus shape in each quadrant as compared with the symmetrical shape of the convex hull in each quadrant. Quadrants are defined by the major and minor axes as shown in Figure 4. |
| C Color |
Color saturation variation | Mean color saturation across the four quadrants of the nevi, with the quadrants defined by the major and minor axes as shown in Figure 4. |
| Color intensity variation | Mean color intensity across the four quadrants of the nevi, with the quadrants defined by the major and minor axes as shown in Figure 4. | |
| D Diameter (size) |
Area | A measure of pixels within the border. |
| Diameter | The extent of the nevus in the direction of the major axis of an ellipse that has the same second moments as the nevus; illustrated in Figure 4, with the nevus shown in white and a rectangle enclosing the nevus that has the same major axis as the ellipse. | |
| Perimeter | The distance measured around the border. | |
| E Evolution |
Change | Change of any of the features mentioned earlier computed as the difference between the original or older feature value and the newer feature values. |
| Percent change | Change normalized by the original feature value. |
Description of validator application
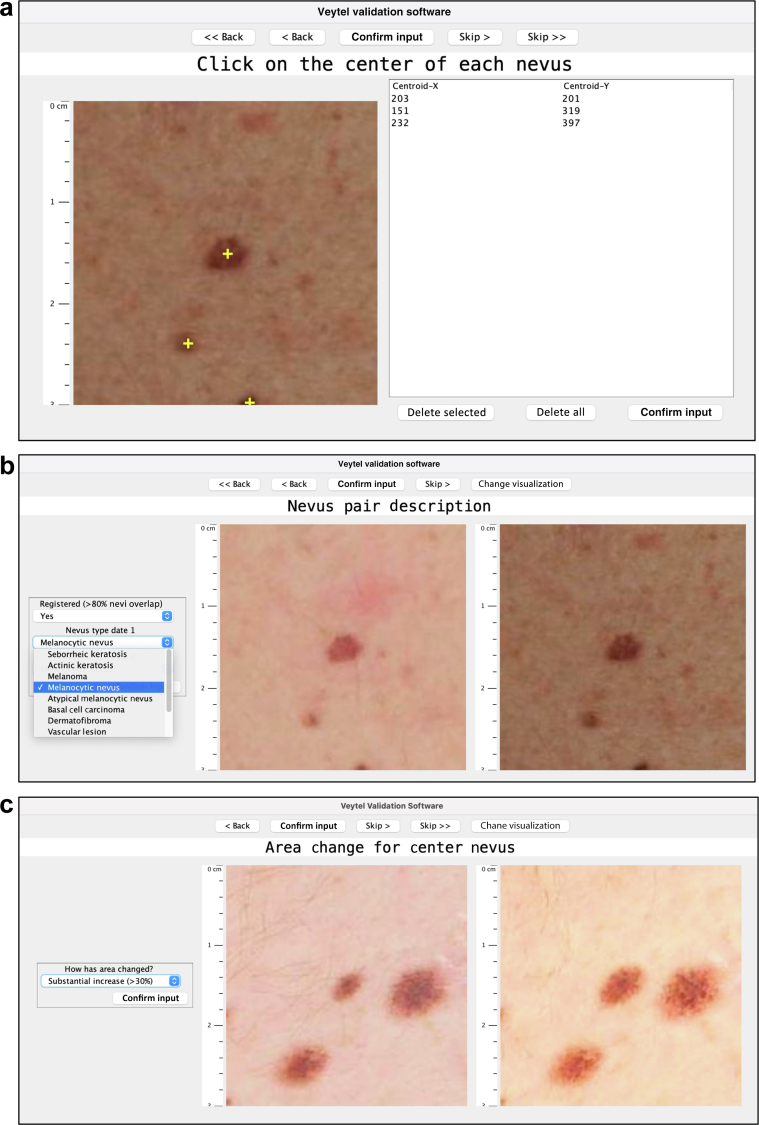
The Validator is an Extensible Markup Language‒based image evaluation software application that allows clinicians to assess features of the DermaViz algorithms and classify and characterize lesions detected by DermaViz. It is highly configurable, allowing the specific prompts and responses to be modified on the basis of the objectives of the study for which it is used. In this study, there are three sections, as illustrated in Figure 5: (i) nevus manual identification (clinician clicks on the center of each nevus to verify the accuracy of software lesion identification), (ii) nevus pair description (clinician assesses the accuracy of lesion registration between time points and identifies the lesion at each time point through options on a drop-down menu), and (iii) area change for the center nevus (clinician enters a subjective opinion of size change along a prespecified scale through a drop-down menu).
Figure 5.
Demonstration of Validator application. (a) Nevus manual identification. (b) Nevus pair description. (c) Area change for center nevus. Images are screenshots from the clinician-facing portion of the Validator application, taken by a clinical reviewer.
The Validator application runs on Mac and Windows computers, with a separate installer for each. On software startup, there is an option to log in as a known user (named in advance in the software) or as an unknown user (delineated by a user number). Upon completion of the questions, the responses are electronically submitted to Veytel (Pittsburgh, PA) for aggregation and analysis.
Characteristics of posterior truncal photographs used in the pilot study
Serial posterior truncal photographs from patients with multiple atypical nevi and a history of melanoma were obtained from a pre-existing image and nevus biobanking protocol database at UPMC Hillman Cancer Center (Pittsburgh, PA). The original images were taken under protocol UPCI 96-099 (institutional review board number REN18100233/IRB970186 approved by the University of Pittsburgh Institutional Review Board) using a single Nikon D700 camera and a standardized background and automatic focus and exposure settings. They were stored using Philips’ iSITE PACS (Philips, Amsterdam, The Netherlands). Patients provided written informed consent for the use of the deidentified images in future research. The images were stripped of all identifiers other than a study-specific identification code and sent to Veytel. Preprocessing consisted only of cropping some images to remove the arms; the images were then analyzed with DermaViz software.
For inclusion in the study, the patients’ images had to meet the following criteria: (i) the number of nevi/lesions imaged in the photos was adequate for registration (roughly greater than 20), (ii) moderate posture changes (not severe), (iii) similar cropping (i.e., including/excluding shoulders), and (iv) an interval between imaging dates <6 years.
For the mobile photographs included in the study, images were acquired using an iPhone 11 (Apple, Cupertino, CA). A volunteer with multiple cutaneous nevi was recruited by Veytel and signed consent forms for both photography and publication of images. Neither the recruitment of the volunteer nor the photography was connected with clinical care; however, the processed images were provided to the patient to share with their healthcare providers.
Change assessment in posterior truncal images
Clinicians were asked to review paired serial posterior truncal images presented as Microsoft PowerPoint slides that could be toggled back and forth between dates. The clinicians first reviewed the set of unmodified photographs and then, in a separate session, reviewed the same images that had been registered and aligned by DermaViz. There was no size threshold for lesions to be included in this portion of the study.
Assessment of sensitivity
Clinicians were shown randomly chosen subfields of the posterior truncal images in the Validator application and asked to click on all pigmented lesions of interest in the images, which marked them with crosshairs and recorded the corresponding coordinates (Figure 5a). The software then determined whether the crosshairs were within the segmentation boundaries of a lesion that had been identified by DermaViz. This allowed the identification of true positives (defined as lesions for which both clinicians and DermaViz identified a nevus), false negatives (defined as lesions for which clinicians but not DermaViz identified a distinct nevus), and non-nevus lesions (defined as lesions identified by DermaViz that were not identified as nevi by clinicians). Sensitivity was calculated as (true positives)/(true positives + false negatives).
Assessment of the accuracy of registration and nevus characterization
For the first method of assessing the quality of registration, reviewers who completed the change assessment in posterior truncal images exercise with aligned PowerPoint slides were asked to comment on whether each image pair had approximately >90% of the lesions accurately registered as well as to comment subjectively on any issues with registration.
For the second method of assessing the quality of registration, a subset of lesions identified by DermaViz was selected for clinician review on the basis of the following criteria: (i) all lesions over 4 mm in diameter; (ii) lesions with visibly apparent changes in paired images as determined by staff at Veytel; and (iii) the largest remaining lesion in each quadrant of the posterior truncal image pair if lesions are present, excluding nevi near the edge of the images.
Clinicians were shown image pairs of randomly chosen lesions photographed on different dates in the Validator application and asked whether the serial image pairs were registered (Figure 5b). Registered was defined in the following way: if there is a change, at least 90% of the area of the smaller of the nevus pair is contained within the larger of the lesion pair. If there is no change, then the lesion pairs overlap by at least 90%. When the lesion pair is not correctly registered, the earlier and later date lesion do not align, that is, there is <90% overlap of the smaller lesion with the larger lesion. Two visualization options were offered: a view with the two time points shown side by side and a toggle view where the aligned images could be toggled back and forth between time points. This section of the Validator also allowed clinicians to classify the lesions by type from a drop-down menu.
Assessment of the accuracy of size change
The first method of assessing the accuracy of size change used the Validator application, for which a subset of lesions was selected for further analysis according to the three criteria specified in the previous section. Clinicians were shown sequential zoomed-in views of isolated lesions, which were chosen at random (Figure 5c). Two visualization options were offered: a view with the two time points shown side by side and a toggle view where the overlaid images could be toggled back and forth between time points. Clinicians were asked to approximately categorize the change observed into specific bins: disappeared (100%), substantial decrease (>30%), moderate decrease (15‒30%), no/minimal change (<15%), moderate increase (15‒30%), and a substantial increase (>30%). The results were compared between observers and with the DermaViz quantitative assessments of diameter and area.
Statistical methods
For the number of new/increased lesions, a linear mixed model was used to study its association with image registration. For the variable detected, which is defined as 1 if a patient had new/increased lesions detected and as 0 otherwise, a linear mixed-effect logistic regression model was used to study its association with image registration. In both of the two mixed models mentioned earlier, the registration (=1 for a registered image, =0 for an unregistered image) was a fixed effect, and patient and clinician were two random effects. The first model was analyzed with Proc Mixed in SAS software (SAS Institute, Cary, NC). The second model was analyzed with SAS Proc GLIMMIX.
For the calculation of CIs for sensitivity and the false negative rate, we used the Clopper‒Pearson interval, which is based on the cumulative probability of the binomial distribution (Clopper and Pearson, 1934).
For assessment of size change, Cohen’s kappa was used to compute kappa values between each individual rater (humans 1‒3 and DermaViz) using both linear and quadratic weights as well as 95% CIs for each comparison. Analysis was performed in Strata (StataCorp, College Station, TX) using the kappaetc program written by Daniel Klein and available through the Boston College Statistical Software Components archive.
Data availability statement
The deidentified posterior truncal images used in this study have been made publicly available and can be found at https://doi.org/10.34970/630662, hosted at the International Skin Imaging Collaboration Archive (Kirkwood, 2022). Requests for additional data related to this study should be addressed to the corresponding author.
ORCIDs
William F. Maguire: http://orcid.org/0000-0002-3288-8863
Paul H. Haley: http://orcid.org/0000-0001-7530-236X
Catherine M. Dietz: http://orcid.org/0000-0002-7672-7973
Mike Hoffelder: http://orcid.org/0000-0002-5220-4756
Clara S. Brandt: http://orcid.org/0000-0003-2097-3005
Robin Joyce: http://orcid.org/0000-0001-5077-087X
Georgia Fitzgerald: http://orcid.org/0000-0003-3180-0839
Christopher Minnier: http://orcid.org/0000-0002-0499-7296
Cindy Sander: http://orcid.org/0000-0002-4136-1342
Laura K. Ferris: http://orcid.org/0000-0002-0598-5182
Gyorgy Paragh: http://orcid.org/0000-0002-6612-9267
Joshua Arbesman: http://orcid.org/0000-0002-6601-7580
Hong Wang: http://orcid.org/0000-0003-0477-2908
Kevin J. Mitchell: http://orcid.org/0000-0002-1870-5522
Ellen K. Hughes: http://orcid.org/0000-0001-7253-4552
John M. Kirkwood: http://orcid.org/0000-0002-3570-4476
Conflict of Interest
PHH, CMD, MH, KJM, and EKH are employees of Veytel. At the time of the study, CSB, RJ, and GF were interns for Veytel. LKF is an investigator for DermTech and Skin Analytics and is a consultant for DermTech. JMK reports consulting for Applied Clinical Intelligence, Amgen, Ankyra Therapeutics, Axio Research/Instil Bio, Becker Pharmaceutical Consulting, Bristol Myers Squibb, Checkmate Pharmaceuticals, DermTech, Fenix Group International, Harbour BioMed, Immunocore LLC, Intellisphere, LLC/Cancer Network, Iovance Biotherapeutics, IQVIA, Istari Oncology, Merck, Millennium Pharmaceuticals/Takeda Pharmaceutical, Natera, Novartis Pharmaceuticals, OncoCyte, OncoSec Medical, Pfizer, Replimune, Scopus BioPharma, and SR One Capital Management and grant support to Institution from Amgen, Bristol Myers Squibb, Castle Biosciences, Checkmate Pharmaceuticals, Harbour BioMed, Immvira Pharma, Immunocore LLC, Iovance Biotherapeutics, Merck, Novartis Pharmaceuticals, Schering-Plough, Takeda, and Verastem. The remaining authors state no conflicts of interest.
Acknowledgments
This research was supported in part by philanthropic support from Making Melanoma History and the Building Bridges effort of the Melanoma Center as well as from the Veytel Research Fund. Veytel is self-funded, and Veytel authors provided the funding for their involvement in this research.
Author Contribution
Conceptualization: WFM, PHH, CMD, MH, KJM, EKH, JMK; Data Curation: WFM, KJM, EKH; Formal Analysis: WFM, PHH, CMD, MH, HW, KJM, EKH; Funding Acquisition: EKH, JMK; Investigation: CS, JMK; Methodology: WFM, CMD, HW, KJM, EKH, JMK; Project Administration; WFM, CS, EKH; Resources: EKH, JMK; Software: PHH, CMD, MH, CSB, RJ, GF, KJM, EKH; Supervision: EKH, KJM, JMK; Validation: WFM, CM, LKF, GP, JA, JMK; Visualization: WFM, EKH; Writing - Original Draft Preparation: WFM, EKH, JMK; Writing - Review and Editing: WFM, PHH, CMD, MH, CSB, RJ, GF, CM, CS, LKF, GP, JA, HW, KJM, EKH, JMK
Disclaimer
University of Pittsburgh Medical Center has no interest in Veytel. No funds changed hands between Veytel and researchers at the University of Pittsburgh/University of Pittsburgh Medical Center in either direction.
accepted manuscript published online XXX; corrected proof published online XXX
Footnotes
Cite this article as: JID Innovations 2022;X:100181
Supplementary material is linked to the online version of the paper at www.jidonline.org, and at https://doi.org/10.1016/j.xjidi.2023.100181
Supplementary Materials
Paired Nikon D700 images of the posterior trunk of a representative patient before and after registration by DermaViz.
Paired iPhone 11 images of the posterior trunk of a volunteer before and after registration by DermaViz.
Paired Nikon D700 images of the posterior trunk of a second representative patient before and after registration by DermaViz.
References
- Banky J.P., Kelly J.W., English D.R., Yeatman J.M., Dowling J.P. Incidence of new and changed nevi and melanomas detected using baseline images and dermoscopy in patients at high risk for melanoma. Arch Dermatol. 2005;141:998–1006. doi: 10.1001/archderm.141.8.998. [DOI] [PubMed] [Google Scholar]
- Blotta E., Bouchet A., Ballarin V., Pastore J. Enhancement of medical images in HSI color space. J Phys Conf Ser. 2011;332:012041. [Google Scholar]
- Chen A.C., Martin A.J., Choy B., Fernández-Peñas P., Dalziell R.A., McKenzie C.A., et al. A phase 3 randomized trial of nicotinamide for skin-cancer chemoprevention. N Engl J Med. 2015;373:1618–1626. doi: 10.1056/NEJMoa1506197. [DOI] [PubMed] [Google Scholar]
- Clopper C.J., Pearson E.S. The use of confidence or fiducial limits illustrated in the case of the binomial. Biometrika. 1934;26:404–413. [Google Scholar]
- Daneshjou R., Barata C., Betz-Stablein B., Celebi M.E., Codella N., Combalia M., et al. Checklist for evaluation of image-based artificial intelligence reports in dermatology: CLEAR Derm consensus guidelines from the International Skin Imaging Collaboration Artificial Intelligence Working Group. JAMA Dermatol. 2022;158:90–96. doi: 10.1001/jamadermatol.2021.4915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Rosario F., Farahi J.M., Drendel J., Buntinx-Krieg T., Caravaglio J., Domozych R., et al. Performance of a computer-aided digital dermoscopic image analyzer for melanoma detection in 1,076 pigmented skin lesion biopsies. J Am Acad Dermatol. 2018;78:927–934.e6. doi: 10.1016/j.jaad.2017.01.049. [DOI] [PubMed] [Google Scholar]
- Dick V., Sinz C., Mittlböck M., Kittler H., Tschandl P. Accuracy of computer-aided diagnosis of melanoma: a meta-analysis. JAMA Dermatol. 2019;155:1291–1299. doi: 10.1001/jamadermatol.2019.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugonik B., Dugonik A., Marovt M., Golob M. Image quality assessment of digital image capturing devices for melanoma detection. Appl Sci. 2020;10(10):2876. [Google Scholar]
- Finnane A., Curiel-Lewandrowski C., Wimberley G., Caffery L., Katragadda C., Halpern A., et al. Proposed technical guidelines for the acquisition of clinical images of skin-related conditions. JAMA Dermatol. 2017;153:453–457. doi: 10.1001/jamadermatol.2016.6214. [DOI] [PubMed] [Google Scholar]
- Fleiss J.L., Cohen J. The equivalence of weighted kappa and the intraclass correlation coefficient as measures of reliability. Educ Psychol Meas. 1973;33:613–619. [Google Scholar]
- Freeman K., Dinnes J., Chuchu N., Takwoingi Y., Bayliss S.E., Matin R.N., et al. Algorithm based smartphone apps to assess risk of skin cancer in adults: systematic review of diagnostic accuracy studies [published correction appears in BMJ 2020;368:m645] BMJ. 2020;368:m127. doi: 10.1136/bmj.m127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller S.R., Bowen G.M., Tanner B., Florell S.R., Grossman D. Digital dermoscopic monitoring of atypical nevi in patients at risk for melanoma. Dermatol Surg. 2007;33:1198–1206. doi: 10.1111/j.1524-4725.2007.33254.x. discussion 1205‒6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazer A.M., Rigel D.S., Winkelmann R.R., Farberg A.S. Clinical diagnosis of skin cancer: enhancing inspection and early recognition. Dermatol Clin. 2017;35:409–416. doi: 10.1016/j.det.2017.06.001. [DOI] [PubMed] [Google Scholar]
- Guan S.-Y., Wang T.-M., Meng C., Wang J.-C. A review of point feature based medical image registration. Chin J Mech Eng. 2018;31:76. [Google Scholar]
- Guido N., Hagstrom E.L., Ibler E., Carneiro C., Orrell K.A., Kelm R.C., et al. A novel total body digital photography smartphone application designed to detect and monitor skin lesions: a pilot study. J Surg Dermatol. 2018;3 [Google Scholar]
- Haenssle H.A., Fink C., Toberer F., Winkler J., Stolz W., Deinlein T., et al. Man against machine reloaded: performance of a market-approved convolutional neural network in classifying a broad spectrum of skin lesions in comparison with 96 dermatologists working under less artificial conditions. Ann Oncol. 2020;31:137–143. doi: 10.1016/j.annonc.2019.10.013. [DOI] [PubMed] [Google Scholar]
- Hornung A., Steeb T., Wessely A., Brinker T.J., Breakell T., Erdmann M., et al. The value of total body photography for the early detection of melanoma: A systematic review. Int J Environ Res Public Health. 2021;18:1726. doi: 10.3390/ijerph18041726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkwood J.M. Longitudinal overview images of posterior trunks. 2022. https://api.isic-archive.com/collections/168/
- Korotkov K., Quintana J., Puig S., Malvehy J., Garcia R. A new total body scanning system for automatic change detection in multiple pigmented skin lesions. IEEE Trans Med Imaging. 2015;34:317–338. doi: 10.1109/TMI.2014.2357715. [DOI] [PubMed] [Google Scholar]
- Levy J.L., Trelles M.A., Levy A., Besson R. Photography in dermatology: comparison between slides and digital imaging. J Cosmet Dermatol. 2003;2:131–134. doi: 10.1111/j.1473-2130.2004.00081.x. [DOI] [PubMed] [Google Scholar]
- Maguire W.F., Kirkwood J.M. Developing agents for the therapeutic prevention of melanoma: can the assessment of cutaneous precursor lesions help? Future Oncol. 2020;16:413–415. doi: 10.2217/fon-2020-0012. [DOI] [PubMed] [Google Scholar]
- Mar V.J., Chamberlain A.J., Kelly J.W., Murray W.K., Thompson J.F. Clinical practice guidelines for the diagnosis and management of melanoma: melanomas that lack classical clinical features. Med J Aust. 2017;207:348–350. doi: 10.5694/mja17.00123. [DOI] [PubMed] [Google Scholar]
- Matin R.N., Dinnes J. AI-based smartphone apps for risk assessment of skin cancer need more evaluation and better regulation. Br J Cancer. 2021;124:1749–1750. doi: 10.1038/s41416-021-01302-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcgregor B. Automatic registration of images of pigmented skin lesions. Pattern Recognit. 1998;31:805–817. [Google Scholar]
- Mirzaalian H., Lee T.K., Hamarneh G. Skin lesion tracking using structured graphical models. Med Image Anal. 2016;27:84–92. doi: 10.1016/j.media.2015.03.001. [DOI] [PubMed] [Google Scholar]
- Morris J.B., Alfonso S.V., Hernandez N., Fernández M.I. Use of and intentions to use dermoscopy among physicians in the United States. Dermatol Pract Concept. 2017;7:7–16. doi: 10.5826/dpc.0702a02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro F., Escudero-Viñolo M., Bescós J. Accurate segmentation and registration of skin lesion images to evaluate lesion change. IEEE J Biomed Health Inform. 2019;23:501–508. doi: 10.1109/JBHI.2018.2825251. [DOI] [PubMed] [Google Scholar]
- Rotemberg V., Kurtansky N., Betz-Stablein B., Caffery L., Chousakos E., Codella N., et al. A patient-centric dataset of images and metadata for identifying melanomas using clinical context [published correction appears in Sci Data 2021;8:81. Sci Data. 2021;8:34. doi: 10.1038/s41597-021-00815-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider S.L., Kohli I., Hamzavi I.H., Council M.L., Rossi A.M., Ozog D.M. Emerging imaging technologies in dermatology: Part II: Applications and limitations. J Am Acad Dermatol. 2019;80:1121–1131. doi: 10.1016/j.jaad.2018.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shain A.H., Bastian B.C. From melanocytes to melanomas [published correction appears in Nat Rev Cancer 2020;20:355] Nat Rev Cancer. 2016;16:345–358. doi: 10.1038/nrc.2016.37. [DOI] [PubMed] [Google Scholar]
- Surjana D., Halliday G.M., Martin A.J., Moloney F.J., Damian D.L. Oral nicotinamide reduces actinic keratoses in phase II double-blinded randomized controlled trials. J Invest Dermatol. 2012;132:1497–1500. doi: 10.1038/jid.2011.459. [DOI] [PubMed] [Google Scholar]
- Tahata S., Singh S.V., Lin Y., Hahm E.R., Beumer J.H., Christner S.M., et al. Evaluation of biodistribution of sulforaphane after administration of oral broccoli sprout extract in melanoma patients with multiple atypical nevi. Cancer Prev Res (Phila.) 2018;11:429–438. doi: 10.1158/1940-6207.CAPR-17-0268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tondewad M.P.S., Dale M.M.P. Remote sensing image registration methodology: review and discussion. Procedia Comput Sci. 2020;171:2390–2399. [Google Scholar]
- Tschandl P., Codella N., Akay B.N., Argenziano G., Braun R.P., Cabo H., et al. Comparison of the accuracy of human readers versus machine-learning algorithms for pigmented skin lesion classification: an open, web-based, international, diagnostic study. Lancet Oncol. 2019;20:938–947. doi: 10.1016/S1470-2045(19)30333-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanbelle S. A new interpretation of the weighted kappa coefficients. Psychometrika. 2016;81:399–410. doi: 10.1007/s11336-014-9439-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Paired Nikon D700 images of the posterior trunk of a representative patient before and after registration by DermaViz.
Paired iPhone 11 images of the posterior trunk of a volunteer before and after registration by DermaViz.
Paired Nikon D700 images of the posterior trunk of a second representative patient before and after registration by DermaViz.
Data Availability Statement
The deidentified posterior truncal images used in this study have been made publicly available and can be found at https://doi.org/10.34970/630662, hosted at the International Skin Imaging Collaboration Archive (Kirkwood, 2022). Requests for additional data related to this study should be addressed to the corresponding author.