Figure 6.
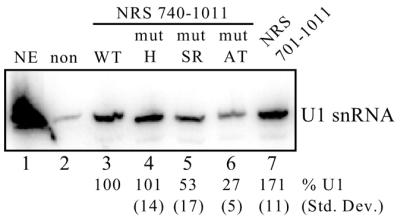
U1 snRNP recruitment to NRS mutants. Equal moles of the indicated biotinylated RNAs were incubated in HeLa nuclear extract and affinity selected with streptavidin–agarose beads. Bound snRNAs were then extracted with phenol/chloroform, separated on a denaturing polyacrylamide gel, electroblotted to a nylon membrane and hybridized to a U1 riboprobe. Non-biotinylated NRS RNA (non) was used to determine background binding to the beads whereas the full-length NRS (nt 701–1011) was used to determine optimal U1 binding (lane 7). To avoid any contributions from the upstream SR protein-binding site, the experimental RNAs lacked the primary SR protein-binding site (nt 740–1011, lanes 3–6). NE, U1 snRNA marker extracted from 3 µl of nuclear extract. Bands were quantitated using a phosphorimager and the average percentage of U1 snRNA selected from three separate experiments is shown below each lane with the corresponding standard deviation. The position of the U1 snRNA is shown on the right. Binding to the wild-type 740–1011 RNA was set at 100% (lane 3).
