Figure 7.
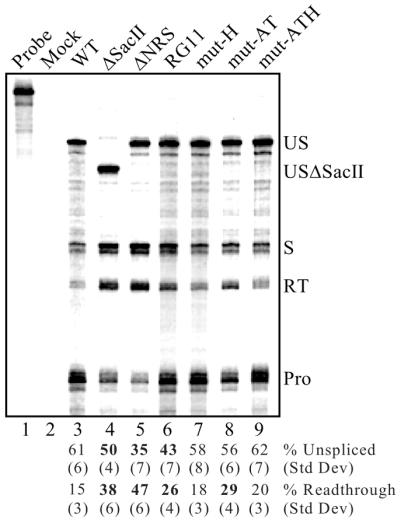
Effect of SR protein- and hnRNP H-specific mutations on splicing and read-through. RNA from CEFs transfected with the corresponding RSV mutants was used for RNase protection assays as described in Figure 6. Protected fragments were separated on a denaturing gel and a phosphorimager was used for visualization and quantitation. Probe, a sample of unprocessed probe; Mock, assay performed using RNA from mock-transfected CEFs. The positions of unspliced (US and USΔSacII), spliced (S), read-through (RT) and processed (Pro) RNAs are shown. Bands were quantitated, normalized for uridine content and the average percentages of unspliced and read-through RNA from six to eight separate transfections and protection assays are shown below the lanes with corresponding standard deviations. Significant deviations (P < 0.002) from wild-type (WT) values are indicated in bold.
